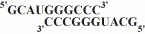Incorporation fidelity of the viral RNA-dependent RNA polymerase: a kinetic, thermodynamic and structural perspective - PubMed (original) (raw)
Review
Incorporation fidelity of the viral RNA-dependent RNA polymerase: a kinetic, thermodynamic and structural perspective
Christian Castro et al. Virus Res. 2005 Feb.
Abstract
Positive-strand RNA viruses exist as a quasi-species due to the incorporation of mutations into the viral genome during replication by the virus-encoded RNA-dependent RNA polymerase (RdRP). Therefore, the RdRP is often described as a low-fidelity enzyme. However, until recently, a complete description of the kinetic, thermodynamic and structural basis for the nucleotide incorporation fidelity of the RdRP has not been available. In this article, we review the following: (i) the steps employed by the RdRP to incorporate a correct nucleotide; (ii) the steps that are employed by the RdRP for nucleotide selection; (iii) the structure-based hypothesis for nucleotide selection; (iv) the impact of sites remote from the active site on polymerase fidelity. Given the recent observation that RNA viruses exist on the threshold of error catastrophe, the studies reviewed herein suggest novel strategies to perturb RdRP fidelity that may lead ultimately to the development of antiviral agents to treat RNA virus infection.
Figures
Fig. 1
Symmetrical primer template substrate (sym/sub) used to study poliovirus polymerase 3Dpol-catalyzed nucleotide incorporation.
Scheme 1
Complete kinetic mechanism for 3Dpol-catalyzed nucleotide incorporation.
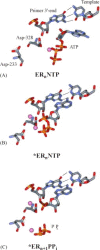
Fig. 2
Structural model for 3Dpol-catalyzed nucleotide incorporation: (A) ground-state binding of metal-complexed nucleotide; (B) reorientation of the triphosphate into the catalytically competent configuration; (C) phosphoryl transfer and pyrophosphate release. While the kinetic mechanism suggests a conformational change prior to pyrophosphate release, kinetic data do not provide any information to permit a molecular description of this step. Images were generated from the model previously described (Gohara et al., 2000). Nucleotide and side chain motions were derived from (Johnson et al., 2003) by approximate rotation and translation movements. Atom colors correspond to the following: red, oxygen; blue, nitrogen; gray, carbon; magenta, Mg2+ or Mn2+. The images were rendered with WebLab Viewer Pro (Accelrys Inc., San Diego, CA). Reproduced with permission from Biochemistry (2004), submitted. © 1998 Am. Chem. Soc.
Scheme 2
Comparison of the conformational-change step and the phosphoryl-transfer step for 3Dpol-catalyzed correct and incorrect nucleotide incorporation in the presence of Mg2+ and Mn2+.
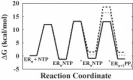
Fig. 3
Comparison of the free energy profile for correct and incorrect 3Dpol-catalyzed nucleotide incorporation in the presence of Mg2+. The free energy profile for correct and incorrect nucleotide incorporation are shown as follows: solid line for AMP incorporation, small dotted line for 2′-dAMP incorporation, and large dotted line for GMP incorporation. The concentrations of the substrates and products used were 2000 μM NTP and 20 μM PP_i_. The free energy for each reaction step was calculated from Δ_G_ = RT [ln (kT/h) − ln (_k_obs,for)], where R = 1.99 cal K−1 mol−1, T = 303 K, k = 3.30 × 10−24 cal K−1, h = 1.58 × 10−34 cal s and k_obs is the first-order rate constant (Arnold et al., 2004). The free energy for each species was calculated from Δ_G = RT [ln (kT/h) − ln (_k_obs,for)] − RT [ln (kT/h) − ln (_k_obs,rev)]. Reproduced with permission from Biochemistry, 2004, submitted. © 1998 Am. Chem. Soc.
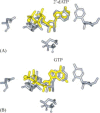
Fig. 4
Structural model for 3Dpol-catalyzed nucleotide incorporation fidelity. Yellow molecules indicate important structural changes: (A) possible conformation of 2′-dATP bound to the NTP-binding pocket; the change here could be caused by the different sugar pucker. (B) Possible conformation of GTP bound to the NTP-binding pocket; the change here could be caused by the non-planar G:U basepair. Reproduced with permission from Biochemistry, 2004, submitted. © 1998 Am. Chem. Soc.
Fig. 5
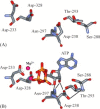
Nucleotide-binding pocket of 3Dpol: (A) residues located in the NTP-binding pocket as observed in the unliganded structure of 3Dpol (Hansen et al., 1997); Asp-233 and Asp-238 are from structural motif A; Ser-288, Thr-293, and Asn-297 are from motif B; Asp-328 is from motif C. (B) Model for interaction of 3Dpol with bound nucleotide (Gohara et al., 2004); ATP and metal ions required for catalysis are labeled. In this model, the side chains for Asp-233 and Asp-238 have been rotated to permit interactions with ATP. Asp-238, Ser-288 and Thr-293 have been positioned to interact. The image was created by using the program WebLab Viewer (Molecular Simulations Inc., San Diego, CA). Reproduced with permission from Biochemistry, 2004, submitted. © 1998 Am. Chem. Soc.
Fig. 6
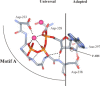
A conserved mechanism for linking binding of a correct nucleotide to the efficiency of phosphoryl transfer. The nucleotide-binding pocket of all nucleic acid polymerases with a canonical “palm”-based active site is highly conserved. The site can be divided into two parts: a region that has “universal” interactions mediated by conserved structural motif A that organizes the metals and triphosphate for catalysis and a region that has “adapted” interactions mediated by conserved structural motif B that dictate whether ribo- or 2′-deoxribonucleotides will be utilized. In the classical polymerase, there is a motif A residue located in the sugar-binding pocket capable of interacting with motif B residue(s) involved in sugar selection. This motif A residue in other polymerases could represent the link between the nature of the bound nucleotide (correct vs. incorrect) to the efficiency of phosphoryl transfer as described herein for Asp-238 of 3Dpol (Gohara et al., 2004). Reproduced with permission from Biochemistry, 2004, submitted. © 1998 Am. Chem. Soc.
Fig. 7
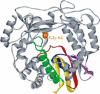
Location of Gly-64 in the structural model of 3Dpol. Model of 3Dpol (complete) based upon sequence and structural homology to rabbit hemorrhagic disease virus 3Dpol (Ng et al., 2002). The conserved structural motifs in the palm subdomain are colored as follows: motif A, red; motif B, green; motif C, yellow; motif D, blue; motif E, purple. van der Waal's projection of Gly-64 (orange). The image was rendered using the program WebLab Viewer Pro (Molecular Simulations Inc., San Diego, CA).
Similar articles
- Molecular dynamics simulations of viral RNA polymerases link conserved and correlated motions of functional elements to fidelity.
Moustafa IM, Shen H, Morton B, Colina CM, Cameron CE. Moustafa IM, et al. J Mol Biol. 2011 Jul 1;410(1):159-81. doi: 10.1016/j.jmb.2011.04.078. Epub 2011 May 7. J Mol Biol. 2011. PMID: 21575642 Free PMC article. - 2'-C-methylated nucleotides terminate virus RNA synthesis by preventing active site closure of the viral RNA-dependent RNA polymerase.
Boehr AK, Arnold JJ, Oh HS, Cameron CE, Boehr DD. Boehr AK, et al. J Biol Chem. 2019 Nov 8;294(45):16897-16907. doi: 10.1074/jbc.RA119.010214. Epub 2019 Oct 1. J Biol Chem. 2019. PMID: 31575662 Free PMC article. - Dynamics: the missing link between structure and function of the viral RNA-dependent RNA polymerase?
Cameron CE, Moustafa IM, Arnold JJ. Cameron CE, et al. Curr Opin Struct Biol. 2009 Dec;19(6):768-74. doi: 10.1016/j.sbi.2009.10.012. Epub 2009 Nov 10. Curr Opin Struct Biol. 2009. PMID: 19910183 Free PMC article. Review. - Homology-Based Identification of a Mutation in the Coronavirus RNA-Dependent RNA Polymerase That Confers Resistance to Multiple Mutagens.
Sexton NR, Smith EC, Blanc H, Vignuzzi M, Peersen OB, Denison MR. Sexton NR, et al. J Virol. 2016 Jul 27;90(16):7415-7428. doi: 10.1128/JVI.00080-16. Print 2016 Aug 15. J Virol. 2016. PMID: 27279608 Free PMC article. - Structure-function relationships among RNA-dependent RNA polymerases.
Ng KK, Arnold JJ, Cameron CE. Ng KK, et al. Curr Top Microbiol Immunol. 2008;320:137-56. doi: 10.1007/978-3-540-75157-1_7. Curr Top Microbiol Immunol. 2008. PMID: 18268843 Free PMC article. Review.
Cited by
- The Pathogenesis and Prevention of Encephalitis due to Human Enterovirus 71.
Bek EJ, McMinn PC. Bek EJ, et al. Curr Infect Dis Rep. 2012 Aug;14(4):397-407. doi: 10.1007/s11908-012-0267-3. Curr Infect Dis Rep. 2012. PMID: 22639066 - Recombination in feline lentiviral genomes during experimental cross-species infection.
Poss M, Idoine A, Ross HA, Terwee JA, VandeWoude S, Rodrigo A. Poss M, et al. Virology. 2007 Mar 1;359(1):146-51. doi: 10.1016/j.virol.2006.08.026. Epub 2006 Oct 12. Virology. 2007. PMID: 17046045 Free PMC article. - Fidelity Characterization of Highly Pathogenic Porcine Reproductive and Respiratory Syndrome Virus and NADC30-like Strain.
Gao X, Bian T, Gao P, Ge X, Zhang Y, Han J, Guo X, Zhou L, Yang H. Gao X, et al. Viruses. 2024 May 16;16(5):797. doi: 10.3390/v16050797. Viruses. 2024. PMID: 38793678 Free PMC article. - SARS-CoV-2 vaccines: What we know, what we can do to improve them and what we could learn from other well-known viruses.
Fiorino S, Carusi A, Hong W, Cernuschi P, Gallo CG, Ferrara E, Maloberti T, Visani M, Lari F, de Biase D, Zippi M. Fiorino S, et al. AIMS Microbiol. 2022 Nov 16;8(4):422-453. doi: 10.3934/microbiol.2022029. eCollection 2022. AIMS Microbiol. 2022. PMID: 36694588 Free PMC article. - Molecular detection, phylogenetic analysis and genetic diversity of recently isolated foot-and-mouth disease virus serotype A African topotype, Genotype IV.
Hassan AM, Zaher MR, Hassanien RT, Abd-El-Moniem MI, Habashi AR, Ibraheem EM, Shahein MA, El Zowalaty ME, Hagag NM. Hassan AM, et al. Virol J. 2022 Jan 3;19(1):1. doi: 10.1186/s12985-021-01693-y. Virol J. 2022. PMID: 34980196 Free PMC article.
References
- Airaksinen A., Pariente N., Menendez-Arias L., Domingo E. Curing of foot-and-mouth disease virus from persistently infected cells by ribavirin involves enhanced mutagenesis. Virology. 2003;311:339–349. - PubMed
- Arnold J.J., Cameron C.E. Poliovirus RNA-dependent RNA polymerase (3Dpol): assembly of stable, elongation-competent complexes by using a symmetrical primer-template substrate (sym/sub) J. Biol. Chem. 2000;275:5329–5336. - PubMed
- Arnold, J.J., Cameron, C.E., 2004a, unpublished observations.
- Arnold J.J., Ghosh S.K., Cameron C.E. Poliovirus RNA-dependent RNA polymerase (3Dpol): divalent cation modulation of primer, template, and nucleotide selection. J. Biol. Chem. 1999;274:37060–37069. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 AI045818/AI/NIAID NIH HHS/United States
- R01 AI045818-07/AI/NIAID NIH HHS/United States
- R01 AI045818-08/AI/NIAID NIH HHS/United States
- AI45818/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Research Materials