Heterodimers of retinoic acid receptors and thyroid hormone receptors display unique combinatorial regulatory properties - PubMed (original) (raw)
Heterodimers of retinoic acid receptors and thyroid hormone receptors display unique combinatorial regulatory properties
Sangho Lee et al. Mol Endocrinol. 2005 Apr.
Abstract
Nuclear receptors are ligand-regulated transcription factors that regulate key aspects of metazoan development, differentiation, and homeostasis. Nuclear receptors recognize target genes by binding to specific DNA recognition sequences, denoted hormone response elements (HREs). Many nuclear receptors can recognize HREs as either homodimers or heterodimers. Retinoid X receptors (RXRs), in particular, serve as important heterodimer partners for many other nuclear receptors, including thyroid hormone receptors (TRs), and RXR/TR heterodimers have been proposed to be the primary mediators of target gene regulation by T3 hormone. Here, we report that the retinoic acid receptors (RARs), a distinct class of nuclear receptors, are also efficient heterodimer partners for TRs. These RAR/TR heterodimers form with similar affinities as RXR/TR heterodimers on an assortment of consensus and natural HREs, and preferentially assemble with the RAR partner 5' of the TR moiety. The corepressor and coactivator recruitment properties of these RAR/TR heterodimers and their transcriptional activities in vivo are distinct from those observed with the corresponding RXR heterodimers. Our studies indicate that RXRs are not unique in their ability to partner with TRs, and that RARs can also serve as robust heterodimer partners and combinatorial regulators of T3-modulated gene expression.
Figures
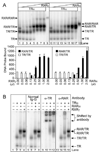
Fig. 1. RARα/TRα Heterodimers Assemble on a DR-4 DNA Response Element
EMSAs were used to determine the ability of TRα, RARα, and RXRγ receptors to bind to DNA as homodimers and heterodimers in the absence of hormone. A, TRα forms heterodimers with both RARα and RXRγ. TRα protein (~40 ng) was incubated with a radiolabeled DR-4 probe together with increasing amounts of RXRγ (~28 ng/µl; lanes 2–8) or RARα (~28 ng/µl; lanes 11–18). Controls included TRα alone (lanes 1 and 10), RXRγ alone (lane 9), and RARα alone (lane 18); equal amounts of nonrecombinant extracts were employed where necessary to keep all assays equivalent. Arrows indicate DNA complexes representing TR monomers (TR), TR homodimers (TR/TR), RAR/TR heterodimers (RAR/TR), and RXR/TR heterodimers (RXR/TR). The amount of radiolabeled DNA probe migrating as TRα homodimer (open bars), RXRγ/TRα heterodimer (filled bars, left panel), or RARα/TRα heterodimer (filled bars, right panel) was quantified below. The mean and
se
of two or more experiments are shown. B, The identity of the various receptor/DNA complexes was confirmed by antibody supershift experiments. EMSAs were performed as in panel A, except the resulting receptor/DNA complexes were incubated with the antisera indicated above each panel before electrophoresis. TRα, RARα, and RXRγ were used individually or in combination (as indicated above the panels) at approximately 30, 14, and 7 ng per assay, respectively. The positions of the various receptor/DNA complexes and the complexes supershifted by antisera are indicated on the right.
Fig. 2. RARα/TRα Heterodimers Form on a Series of Prototypic DNA Response Elements
The ability of RARα and TRα to bind as homodimers and as heterodimers to a variety of DNA response elements was tested using an EMSA protocol as in Fig. 1. Radiolabeled DR-4, DR-5, DIV-6, and INV-0 DNA elements, all composed of AGGTCA half-site repeats, were tested. TRα and RARα proteins, used at 40 and 28 ng respectively, were assayed alone (left and right panels), or as an RARα/TRα mixture (center panels). Where indicated, T3 or ATRA was also included at 1 µM in the EMSA binding buffer. The amount of radiolabeled DNA probe migrating as a TRα homodimer (open bars), RARα homodimer (stippled bars), or RARα/TRα heterodimer (filled bars) under each condition was quantified; the mean and
se
of three experiments are shown.
Fig. 3. The Ability to Form RAR/TR Heterodimers Is Shared by Different Receptor Isotypes
The ability of different RAR and TR isotypes to bind as heterodimers was tested using an EMSA protocol as in Fig. 1. A, Both TRα and TRβ1 can form heterodimers with RARα on a DR-4 element. A fixed amount of RARα (21 ng) was mixed with increasing amounts of either TRα or TRβ (40 ng/µl) as indicated below the panel. The amount of radiolabeled DNA probe migrating as a RAR/TR heterodimer was quantified and is presented (filled bars). B, Three isoforms of RAR form heterodimers with TRα with different efficiencies. TRα (40 ng) and RARα, β, or γ protein preparations (normalized to yield equal homodimer formation when tested alone; ~21 ng for RARα) were assayed alone (left and center panels), or as a RAR/TRα mixture (right panels). DR-4, DR-5, and the F2-DIV-6 element were employed as radiolabeled probes. The amount of radiolabeled DNA probe migrating as a TRα homodimer (open bars), RAR homodimer (stippled bars), or RAR/TRα heterodimer (filled bars) was quantified and is presented. The mean and range of two experiments are shown. No hormone was used in these assays.
Fig. 4. RAR/TR Heterodimers Assemble with a Defined Polarity on DR-4 DNA Response Element
The ability of TRα to bind as a homodimer or as an RARα/TRα heterodimer to DNA elements bearing different half-sites was tested using the EMSA protocol in Fig. 1. A set of radiolabeled DNA elements was created containing a combination of consensus (AGGTCA), RAR-selective (AGTTCA), or TR-selective (AGGACA) half-sites, oriented as a DR-4 or a DR-5, as indicated in each panel (A–F). TRα protein (~40 ng) was incubated with each DNA probe either alone or together with increasing amounts of RARα (~28 ng/µl) as indicated below the panels, and the resulting DNA complexes were resolved by EMSA. The amount of radiolabeled probe migrating as a TRα homodimer (open bars) or as an RARα/TRα heterodimer (filled bars) was quantified as in Fig. 1. RARα homodimers formed in the absence of TRα were also quantified (stippled bars). Comparable results were obtained in repeated studies; a representative experiment is presented. No hormone was used in these assays.
Fig. 5. RAR/TR Heterodimers Bind to Natural DNA Response Elements
TRα, RARα, RARβ, and RXRγ proteins were tested for the ability to bind to different natural and synthetic DNA elements using an EMSA protocol as in Fig. 1. TRα, RARα, and RXRγ were used at approximately 40 ng, 20 ng, and 11 ng per assay, respectively; RARβ was titrated to yield equal homodimer formation as RARα. Radiolabeled DNA probes included a consensus DR-4, a consensus DR-5, the retinoid response element in the RARβ promoter (β-RARE), or the T3-response elements found in the F2-lysozyme promoter (F2-DIV-6), rGH promoter, rMHC promoter, human MyoD promoter, and α-actin promoter. For each probe, receptor preparations were tested individually or in combination as indicated above each panel. The positions of receptor/DNA complexes representing TR monomers, TR homodimers, RAR/TR heterodimers, RXR/TR heterodimers, and RXR/RAR heterodimers are indicated to the left of the panels. Comparable results were obtained in repeated studies; a representative experiment is presented. No hormone was used in these assays.
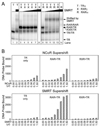
Fig. 6. RAR/TR Heterodimers Recruit SMRT and N-CoR Corepressors
The ability of RARα/TRα and RXRγ/TRα heterodimers to bind to SMRT or N-CoR corepressor was determined using a radiolabeled DR-4 DNA probe and an EMSA supershift protocol. A, PhosphorImager depiction of EMSA. TRα homodimers (lanes 1 and 2), RXRγ homodimers (lane 8), RARα homodimers (lane 14), RXRγ/TRα heterodimers (lanes 3–7), or RARα/TRα heterodimers (lanes 9–13) were allowed to bind to the radiolabeled DR-4 DNA probe as in Fig. 1, but in the presence of differing concentrations of a GST-SMRT construct, as indicated above the panels. The positions of the various receptor/DNA complexes are indicated on the right; an interaction with SMRT results in a supershifted receptor/DNA complex of slower mobility (indicated). TRα, RARα, and RXRγ were used at 40, 11, and 11 ng per assay, respectively; GST-SMRT was used at 200 ng/µl. B, SMRT binds to RARα/TRα heterodimers more efficiently than does N-CoR. TRα only (left panels), RXRγ/TRα heterodimers (right panels), and RARα/TRα heterodimers (center panels) were allowed to bind to a radiolabeled DR-4 DNA probe in the presence of increasing concentrations of either GST-N-CoR (top) or GST-SMRT (bottom); both corepressor preparations were 200 ng protein/µl. The amount of supershifted complex was quantified by PhosphorImager analysis and is presented. Comparable results were obtained in repeated studies; a representative experiment is presented. No hormone was used in these assays.
Fig. 7. Both T3 and ATRA Are Required for Efficient SMRT Release from RARα/TRα Heterodimers
TRα homodimers (left panels), RARα homodimers (right panels), and RARα/TRα heterodimers (center panels) were assembled on the indicated DNA probes in the presence of the GST-SMRT construct as in Fig. 6. T3 alone, ATRA alone, or T3 and ATRA together were added at 1 µM concentrations, and the resulting protein/DNA complexes were resolved by EMSA. The amount of receptor/DNA complex bound by SMRT (i.e. supershifted by GST-SMRT) was quantified for each condition and is presented (cross-hatched bars). TRα, RARα, and GST-SMRT were used at 40, 28, and 200 ng per assay, respectively. The mean and
se
of three experiments are shown.
Fig. 8. Coactivators Are Recruited by RARα/TRα Heterodimers in Response to Cognate Hormones
The ability of various TR, RAR, and RXR complexes to bind ACTR coactivator in response to different hormones was determined. A, TRα homodimers (TR only), RARα homodimers (RAR only), RXRγ homodimers (RXR only), RARα/TRα heterodimers (RAR + TR), and RXRγ/TRα heterodimers (RXR + TR) were assembled on a radiolabeled DR-4 DNA probe in the presence of a GST-ACTR coactivator construct. T3 alone, ATRA alone, 9-cis retinoic acid, or T3 + ATRA was included in the binding assay at 1 µM concentrations as indicated below the panels. The resulting protein/DNA complexes were resolved by EMSA. The amount of radiolabeled probe migrating as a TRα (open bars), RARα, or RXRγ homodimer (stippled bars); as an RAR/TR or RXR/TR heterodimer (filled bars); or as an ACTR supershifted receptor/DNA complex (crosshatched bars) was quantified under each condition and is presented. TRα, RARα, RXRγ, and GST-ACTR were used at 40 ng, 21 ng, 7 ng, and 1 µg per assay respectively. B, The same experiment as in panel A was performed employing a GST-SRC-1 coactivator construct (1 µg per assay). C, The EMSA supershift experiments in A and B were repeated using a competition strategy. RARβ/TRα heterodimers, assembled on a DR-4 DNA probe, were mixed with 2 µg of the SRC-1 construct either alone or with increasing amounts (0.0156 to 1 µg in 2-fold increments) of the ACTR construct; the experiment was performed in the presence of 1 µM T3 or ATRA, as indicated. The amount of RARβ/TRα/DNA complex supershifted into a SRC-1 or into an ACTR complex was determined for each ACTR concentration as for panels A and B. The mean and
se
of three experiments are shown.
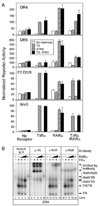
Fig. 9. TRα and RARα Form Heterodimers and Mediate Combinatorial Transcriptional Regulation When Coexpressed in Mammalian Cells
Transient transfections were performed to assay reporter gene regulation and heterodimer formation by RARs and TRs. A, A reporter plasmid (100 ng) bearing the response elements indicated was introduced into CV-1 cells together with 2 ng of an empty pSG5 expression vector (no receptor), 2 ng of pSG5-TRα, 2 ng of pSG5-RARα, or 2 ng each of the combined TRα and RARα expression vectors, as indicated below the panels. A pCH110 β-galactosidase reporter (25 ng) was included in all transfections as an internal control. After transfection, the cells were transferred into fresh media supplemented with ethanol carrier alone (open bars), 500 nM T3 (cross-hatched bars), 500 nM ATRA (stippled bars), or T3 and ATRA together (filled bars). The cells were subsequently lysed and analyzed for luciferase and β-galactosidase activity. Relative luciferase activity (absolute luciferase divided by β-galactosidase activity) is presented for three transfection experiments; the average and SD values are shown. B, COS-1 cells were transfected with empty pSG5 expression vector or the pSG5 vector expressing TRα or RARα. Nuclear extracts were isolated and mixed with radiolabeled DR-4 probe, and the resulting receptor/DNA complexes were resolved by EMSA. Two concentrations of RARα were employed in the mixing experiments with TRα (lanes 3, 4, 8, 9, 13, 14, 18, and 19). Antibody (either normal IgG, anti-TR, anti-RAR, or anti-RXR, as in Fig. 1B) was included in the EMSA binding reactions, indicated above each panel, to confirm the identity of the various receptor/DNA complexes by supershift.
Fig. 10. RAR and TR Function Combinatorially in S. cerevisae
Expression vectors (pG1 or pHG2) for RARβ, TRβ, or RXRγ were introduced, singularly or in the combinations indicated, together with a DR-4 β-galactosidase reporter (Δss-DR4) into the W303 strain of S. cerevisae as previously described (62). The yeast were propagated 12 h in the presence of 5 µM T3, 5 µM ATRA, or equivalent amounts of ethanol carrier, as indicated, harvested, and assayed for β-galactosidase activity and for culture density. Relative β-galactosidase activity (β-galactosidase normalized to culture density) was determined for two independent transformants for each receptor combination; the mean and range are provided. A, Cointroduction of either RARβ or RXRγ enhances reporter gene activation by TRβ1 relative to TRβ1 alone. B, Cointroduction of RARβ and TRβ1 enhances the TRβ1 response to T3, but not the RARβ response to ATRA, relative to the same receptors assayed individually.
Similar articles
- Heterodimeric receptor complexes determine 3,5,3'-triiodothyronine and retinoid signaling specificities.
Hermann T, Hoffmann B, Zhang XK, Tran P, Pfahl M. Hermann T, et al. Mol Endocrinol. 1992 Jul;6(7):1153-62. doi: 10.1210/mend.6.7.1324421. Mol Endocrinol. 1992. PMID: 1324421 - Analysis of the functional role of steroid receptor coactivator-1 in ligand-induced transactivation by thyroid hormone receptor.
Jeyakumar M, Tanen MR, Bagchi MK. Jeyakumar M, et al. Mol Endocrinol. 1997 Jun;11(6):755-67. doi: 10.1210/mend.11.6.0003. Mol Endocrinol. 1997. PMID: 9171239 - The function of retinoid X receptors on negative thyroid hormone response elements.
Takeda T, Nagasawa T, Miyamoto T, Hashizume K, DeGroot LJ. Takeda T, et al. Mol Cell Endocrinol. 1997 Apr 4;128(1-2):85-96. doi: 10.1016/s0303-7207(97)04025-2. Mol Cell Endocrinol. 1997. PMID: 9140079 - Hetero- and homodimeric receptors in thyroid hormone and vitamin A action.
Zhang XK, Pfahl M. Zhang XK, et al. Receptor. 1993 Fall;3(3):183-91. Receptor. 1993. PMID: 8167569 Review. - [Genetic control of the development by retinoic acid].
Mark M, Kastner P, Ghyselinck NB, Krezel W, Dupé V, Chambon P. Mark M, et al. C R Seances Soc Biol Fil. 1997;191(1):77-90. C R Seances Soc Biol Fil. 1997. PMID: 9181129 Review. French.
Cited by
- Targeting Androgen, Thyroid Hormone, and Vitamin A and D Receptors to Treat Prostate Cancer.
Hantusch B, Kenner L, Stanulović VS, Hoogenkamp M, Brown G. Hantusch B, et al. Int J Mol Sci. 2024 Aug 26;25(17):9245. doi: 10.3390/ijms25179245. Int J Mol Sci. 2024. PMID: 39273194 Free PMC article. Review. - A System Biology Approach Reveals New Targets for Human Thyroid Gland Toxicity in Embryos and Adult Individuals.
Oliveira JM, Zenzeluk J, Serrano-Nascimento C, Romano MA, Romano RM. Oliveira JM, et al. Metabolites. 2024 Apr 16;14(4):226. doi: 10.3390/metabo14040226. Metabolites. 2024. PMID: 38668354 Free PMC article. - Subcutaneous adipose tissue alteration in aging process associated with thyroid hormone signaling.
Zhang WN, Zhu H, Ma ZW, Yu J, Yang Y, Lu XB, Lv YF, Wang XD. Zhang WN, et al. BMC Med Genomics. 2023 Aug 25;16(1):202. doi: 10.1186/s12920-023-01641-5. BMC Med Genomics. 2023. PMID: 37626392 Free PMC article. - The Role of Estrogen and Thyroid Hormones in Zebrafish Visual System Function.
Cohen A, Popowitz J, Delbridge-Perry M, Rowe CJ, Connaughton VP. Cohen A, et al. Front Pharmacol. 2022 Feb 28;13:837687. doi: 10.3389/fphar.2022.837687. eCollection 2022. Front Pharmacol. 2022. PMID: 35295340 Free PMC article. Review. - Retinoic acid signaling is critical during the totipotency window in early mammalian development.
Iturbide A, Ruiz Tejada Segura ML, Noll C, Schorpp K, Rothenaigner I, Ruiz-Morales ER, Lubatti G, Agami A, Hadian K, Scialdone A, Torres-Padilla ME. Iturbide A, et al. Nat Struct Mol Biol. 2021 Jun;28(6):521-532. doi: 10.1038/s41594-021-00590-w. Epub 2021 May 27. Nat Struct Mol Biol. 2021. PMID: 34045724 Free PMC article.
References
- Apriletti JW, Ribeiro RC, Wagner RL, Feng W, Webb P, Kushner PJ, West BL, Nilsson S, Scanlan TS, Fletterick RJ, Baxter JD. Molecular and structural biology of thyroid hormone receptors. Clin Exp Pharmacol Physiol Suppl. 1998;25:S2–S11. - PubMed
- Beato M, Klug J. Steroid hormone receptors: an update. Human Reprod Update. 2000;6:225–236. - PubMed
- Chambon P. A decade of molecular biology of retinoic acid receptors. FASEB J. 1996;10:940–954. - PubMed
- Zhang J, Lazar MA. The mechanism of action of thyroid hormones. Annu Rev Physiol. 2000;62:439–466. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R37 CA053394/CA/NCI NIH HHS/United States
- R01 DK053528-07/DK/NIDDK NIH HHS/United States
- R01 DK053528/DK/NIDDK NIH HHS/United States
- R37 CA53394/CA/NCI NIH HHS/United States
- R01 DK053528-10/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Miscellaneous