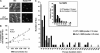A genetic screen for top3 suppressors in Saccharomyces cerevisiae identifies SHU1, SHU2, PSY3 and CSM2: four genes involved in error-free DNA repair - PubMed (original) (raw)
A genetic screen for top3 suppressors in Saccharomyces cerevisiae identifies SHU1, SHU2, PSY3 and CSM2: four genes involved in error-free DNA repair
Erika Shor et al. Genetics. 2005 Mar.
Abstract
Helicases of the RecQ family and topoisomerase III are evolutionarily conserved proteins important for maintenance of genome stability. In Saccharomyces cerevisiae, loss of the TOP3 gene, encoding topoisomerase III, results in a phenotype of slow growth, DNA damage sensitivity, meiotic defects, and hyperrecombination. The sole RecQ helicase in budding yeast, Sgs1, interacts with Top3 both physically and genetically, and the two proteins are thought to act in concert in vivo. Much recent genetic and biochemical evidence points to the role of RecQ helicases and topoisomerase III in regulating homologous recombination (HR) during DNA replication. Previously, we found that mutations in HR genes partially suppress top3 slow growth. Here, we describe the analysis of four additional mutational suppressors of top3 defects: shu1, shu2, psy3, and csm2. These genes belong to one epistasis group and their protein products interact with each other, strongly suggesting that they function as a complex in vivo. Their mutant phenotype indicates that they are important for error-free repair of spontaneous and induced DNA lesions, protecting the genome from mutation. These mutants exhibit an epistatic relationship with rad52 and show altered dynamics of Rad52-YFP foci, suggesting a role for these proteins in recombinational repair.
Figures
Figure 1.—
Genetic interactions among sgs1, top3, and the shu mutants. All the mutations shown are deletions of the respective genes. The haploid strains are meiotic segregants of diploids ESD50, ESD51, W4105, and W4173. For the DNA damage sensitivity assays in B–D, 10-fold serial dilutions of overnight cultures were spotted onto YPD medium containing the indicated drugs. (A) Deletion of SHU1, SHU2, PSY3, or CSM2 suppresses _top3_Δ slow growth. Doubling times in rich medium are shown. (B) Deletion of SHU1, SHU2, PSY3, or CSM2 suppresses _top3_Δ sensitivity to HU and MMS. (C) Deletion of SHU1, SHU2, PSY3, or CSM2 suppresses _sgs1_Δ HU sensitivity. (D) SHU1, SHU2, PSY3, and CSM2 are in one epistasis group for MMS resistance. The quadruple deletion mutant is no more MMS sensitive than any of the single mutants.
Figure 2.—
Effects of _shu1_Δ on the formation of Rad52-YFP foci in the _top3_Δ and _sgs1_Δ mutants. The haploid strains for these experiments were generated from diploids ESD35 and ESD46. (A) The _top3_Δ strain exhibits an increase in Rad52-YFP foci compared to the wild-type strain. (B) _shu1_Δ partially suppresses increased Rad52 foci in the top3 mutant. The graph shows the percentage of cells in G1 (i.e., unbudded cells) or in S/G2/M (i.e., all budded cells) that contain Rad52-YFP foci in the indicated strains. The inset shows the cell cycle distribution of the same four strains. The graph represents data from at least 300 cells of each genotype. (C) _shu1_Δ partially suppresses increased Rad52-YFP foci in the _sgs1_Δ mutant after HU treatment. The graph shows the percentage of HU-treated and untreated cells in G1 or S/G2/M that contain Rad52-YFP foci. The two insets show the cell cycle distribution of untreated cells as well as of cells treated with 100 m
m
HU for 100 min. Between 150 and 300 cells were examined for every strain for either condition (HU or no HU) during this experiment.
Figure 3.—
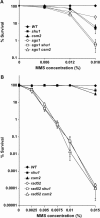
The SHU genes are in the RAD52 epistasis group for MMS resistance. All mutations shown are gene deletions. When comparing the data shown in A to those in B, note the difference in MMS concentrations. (A) Survival curves showing that SGS1 and the SHU genes are in different epistasis groups for MMS resistance. Deletion of SGS1 and either SHU1 or CSM2 results in additional MMS sensitivity compared to any of the single mutants. (B) Survival curves showing that _rad52_Δ _shu1_Δ and _rad52_Δ _csm2_Δ double mutants are no more MMS sensitive than the _rad52_Δ single mutant. At the highest MMS concentration used in these experiments (0.015%), deletion of SHU1 or CSM2 results in a two- to threefold decrease in survival compared to the wild-type strain.
Figure 4.—
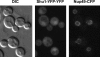
Shu1-YFP-YFP exhibits predominantly nuclear localization. Differential interference contrast (DIC), YFP, and CFP images of a representative field of cells expressing Shu1-YFP-YFP and Nup49-CFP proteins are shown. Nup49 is a nuclear envelope protein. The Shu1-YFP-YFP fluorescent signal is strongest within the Nup49-CFP-defined nuclear boundaries.
Figure 5.—
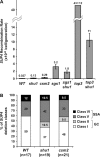
Effect of deletion of SHU1 or CSM2 on recombination at the _SUP4_-o locus in an otherwise wild-type background, as well as in _sgs1_Δ and _top3_Δ mutants. Diploids ESD45, ESD47, and ESD48 produced the haploid strains shown. (A) _shu1_Δ and _csm2_Δ mutants show a three- to sevenfold increase in the _SUP4_-o recombination rate. Deletion of SHU1 partially suppresses the hyperrecombination phenotype of _sgs1_Δ and _top3_Δ mutants. (B) _shu1_Δ and _csm2_Δ mutants do not affect SUP4 deletion class distribution. n, number of independently generated SUP4 recombinants tested for each strain.
Figure 6.—
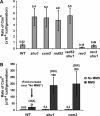
Deletion of SHU1 or CSM2 results in a mutator phenotype. The forward mutation rates of the CAN1 gene are shown. The strains shown are meiotic segregants of diploids ESD41, ESD44, and ESD49. (A) Spontaneous CAN1 mutation rates. rad52 is epistatic to shu1 and csm2: CAN1 mutation rate in the _rad52_Δ _shu1_Δ and _rad52_Δ _csm2_Δ double mutants is similar to that in the single mutants. This graph also shows that the mutator phenotype of _shu1_Δ and _csm2_Δ mutants is dependent on the REV3 gene. (B) MMS-induced CAN1 mutation rates. Treatment with 0.01% MMS stimulates the CAN1 mutation rate ∼35-fold in the wild-type and the mutant strains.
Figure 7.—
Deletion of SHU1 alters Rad52-YFP focus dynamics after DNA damage by MMS. (A) Deletion of SHU1 results in an increase in Rad52-YFP foci upon MMS treatment. Representative samples of wild-type and _shu1_Δ cells treated with 0.05% MMS are shown. (B) The graph shows the response of the wild-type strain and the _shu1_Δ mutant to increasing concentrations of MMS. The percentages of S/G2/M (i.e., all budded) cells containing Rad52-YFP foci are plotted. Fluorescent images were captured after 100 min of growth in the presence of MMS. At least 150 cells were analyzed for each strain under each condition. (C) Time-lapse analyses of Rad52-YFP foci in cells exposed to 0.01% MMS for 100 min. Duration of individual foci is plotted on the _x_-axis vs. the number of foci that lasted for that amount of time. While images of cells were captured every 5 min, the data are plotted in 10-min intervals to reduce complexity. Thus, in the graph, the bars over the 10-min mark correspond to the number of foci that lasted <10 min, the bars over the 20-min mark correspond to the number of foci that lasted between 10 and 20 min, etc. The median duration of Rad52-YFP foci is increased from 15 min in the wild-type strain to 25 min in the _shu1_Δ mutant. The inset shows the data from untreated cells, where the median duration of Rad52-YFP foci is unchanged by _shu1_Δ mutation.
Similar articles
- Mutations in homologous recombination genes rescue top3 slow growth in Saccharomyces cerevisiae.
Shor E, Gangloff S, Wagner M, Weinstein J, Price G, Rothstein R. Shor E, et al. Genetics. 2002 Oct;162(2):647-62. doi: 10.1093/genetics/162.2.647. Genetics. 2002. PMID: 12399378 Free PMC article. - The yeast Shu complex couples error-free post-replication repair to homologous recombination.
Ball LG, Zhang K, Cobb JA, Boone C, Xiao W. Ball LG, et al. Mol Microbiol. 2009 Jul;73(1):89-102. doi: 10.1111/j.1365-2958.2009.06748.x. Epub 2009 Jun 1. Mol Microbiol. 2009. PMID: 19496932 - The absence of Top3 reveals an interaction between the Sgs1 and Pif1 DNA helicases in Saccharomyces cerevisiae.
Wagner M, Price G, Rothstein R. Wagner M, et al. Genetics. 2006 Oct;174(2):555-73. doi: 10.1534/genetics.104.036905. Epub 2006 Jul 2. Genetics. 2006. PMID: 16816432 Free PMC article. - [Functional analysis of yeast homologue gene associated with human DNA helicase causative syndromes].
Miyajima A. Miyajima A. Kokuritsu Iyakuhin Shokuhin Eisei Kenkyusho Hokoku. 2002;(120):53-74. Kokuritsu Iyakuhin Shokuhin Eisei Kenkyusho Hokoku. 2002. PMID: 12638184 Review. Japanese. - DNA sequence analysis of spontaneous mutagenesis in Saccharomyces cerevisiae.
Kunz BA, Ramachandran K, Vonarx EJ. Kunz BA, et al. Genetics. 1998 Apr;148(4):1491-505. doi: 10.1093/genetics/148.4.1491. Genetics. 1998. PMID: 9560369 Free PMC article. Review.
Cited by
- Replication-Associated Recombinational Repair: Lessons from Budding Yeast.
Bonner JN, Zhao X. Bonner JN, et al. Genes (Basel). 2016 Aug 17;7(8):48. doi: 10.3390/genes7080048. Genes (Basel). 2016. PMID: 27548223 Free PMC article. Review. - Homologous recombination and its regulation.
Krejci L, Altmannova V, Spirek M, Zhao X. Krejci L, et al. Nucleic Acids Res. 2012 Jul;40(13):5795-818. doi: 10.1093/nar/gks270. Epub 2012 Mar 30. Nucleic Acids Res. 2012. PMID: 22467216 Free PMC article. Review. - Genome-wide requirements for resistance to functionally distinct DNA-damaging agents.
Lee W, St Onge RP, Proctor M, Flaherty P, Jordan MI, Arkin AP, Davis RW, Nislow C, Giaever G. Lee W, et al. PLoS Genet. 2005 Aug;1(2):e24. doi: 10.1371/journal.pgen.0010024. Epub 2005 Aug 19. PLoS Genet. 2005. PMID: 16121259 Free PMC article. - Replication blocking lesions present a unique substrate for homologous recombination.
Ward JD, Barber LJ, Petalcorin MI, Yanowitz J, Boulton SJ. Ward JD, et al. EMBO J. 2007 Jul 25;26(14):3384-96. doi: 10.1038/sj.emboj.7601766. Epub 2007 Jul 5. EMBO J. 2007. PMID: 17611606 Free PMC article. - Regulation of RAD51 at the Transcriptional and Functional Levels: What Prospects for Cancer Therapy?
Orhan E, Velazquez C, Tabet I, Sardet C, Theillet C. Orhan E, et al. Cancers (Basel). 2021 Jun 11;13(12):2930. doi: 10.3390/cancers13122930. Cancers (Basel). 2021. PMID: 34208195 Free PMC article. Review.
References
- Akada, R., J. Yamamoto and I. Yamashita, 1997. Screening and identification of yeast sequences that cause growth inhibition when overexpressed. Mol. Gen. Genet. 254: 267–274. - PubMed
- Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman et al., 1998 Current Protocols in Molecular Biology. John Wiley & Sons, New York.
- Barbour, L., and W. Xiao, 2003. Regulation of alternative replication bypass pathways at stalled replication forks and its effects on genome stability: a yeast model. Mutat. Res. 532: 137–155. - PubMed
- Bennett, R. J., J. A. Sharp and J. C. Wang, 1998. Purification and characterization of the Sgs1 DNA helicase activity of Saccharomyces cerevisiae. J. Biol. Chem. 273: 9644–9650. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials