Rapid concerted evolution of nuclear ribosomal DNA in two Tragopogon allopolyploids of recent and recurrent origin - PubMed (original) (raw)
Comparative Study
Rapid concerted evolution of nuclear ribosomal DNA in two Tragopogon allopolyploids of recent and recurrent origin
A Kovarik et al. Genetics. 2005 Feb.
Abstract
We investigated concerted evolution of rRNA genes in multiple populations of Tragopogon mirus and T. miscellus, two allotetraploids that formed recurrently within the last 80 years following the introduction of three diploids (T. dubius, T. pratensis, and T. porrifolius) from Europe to North America. Using the earliest herbarium specimens of the allotetraploids (1949 and 1953) to represent the genomic condition near the time of polyploidization, we found that the parental rDNA repeats were inherited in roughly equal numbers. In contrast, in most present-day populations of both tetraploids, the rDNA of T. dubius origin is reduced and may occupy as little as 5% of total rDNA in some individuals. However, in two populations of T. mirus the repeats of T. dubius origin outnumber the repeats of the second diploid parent (T. porrifolius), indicating bidirectional concerted evolution within a single species. In plants of T. miscellus having a low rDNA contribution from T. dubius, the rDNA of T. dubius was nonetheless expressed. We have apparently caught homogenization of rDNA repeats (concerted evolution) in the act, although it has not proceeded to completion in any allopolyploid population yet examined.
Figures
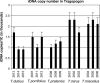
Figure 1.—
Copy numbers of rDNA units in Tragopogon haploid genomes. Quantification was performed by slot-blot analysis using the 26S rDNA hybridization probe. The signal in allotetraploids was nearly the same as in diploids. The exact copy numbers were calculated from known nuclear content for each species (P
ires
et al. 2004), considering ∼10 kb as the length of rDNA operon. Note that copy numbers in tetraploids were slightly less than double those in respective diploid progenitors.
Figure 2.—
SSCP analysis of the ITS1 region in allotetraploid species of Tragopogon and their diploid parents. The amplified region is shown in Figure 3A. The high-resolution gel resolved the profiles in the parental species. Two to three bands were visible in T. dubius, two bands in T. pratensis, and a single band in T. porrifolius under the conditions used. The parental bands are found in all lanes loaded with DNA from the tetraploid species. Note the significant reduction of _T. dubius_-specific bands in the populations of the two allotetraploids, with the exception of T. mirus 2602 (comparable results for all populations were also obtained with cloning and Southern blots).
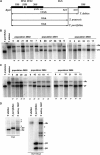
Figure 3.—
Southern blot hybridization and clone analysis of the ITS1 region. (A) Restriction enzyme maps of the major T. dubius, T. pratensis, and T. porrifolius rDNA units. The position of probe hybridization is indicated by a thick line. The sizes of hybridization fragments are indicated. 18S and 5.8S are genes coding for rRNA molecules. Distances are approximately to scale. Thick arrows indicate positions of 18Sfor and 5.8Srev primers. (B) Southern blot hybridization of genomic DNAs showing variability in the parental ITS1 families content among the populations of T. mirus. The DNAs were digested with the _Bst_NI restriction enzyme and hybridized with the ITS1 probe. The diagnostic fragments representing T. dubius (du) and T. porrifolius (po) families are indicated. Sample 2603-33 is shown after longer exposure to reveal a particularly faint ∼500-bp _T. dubius_-specific band in this plant. Numbers in italics below the tracts indicate relative proportions of the _T. dubius_-specific band expressed as a percentage of total signal in tract (also for C and D). (C) Southern blot hybridization of genomic DNAs showing variation in the ITS1 families among populations of T. miscellus. The digestion and hybridization conditions were the same as in B. The diagnostic fragments representing T. dubius (du) and T. pratensis (pr) families are indicated. (D) Analysis of unit inheritance in unusual 2603-33 plants. The sibling plants were grown from seeds collected from different heads (A–C). The Southern blot hybridization and probe was the same as in B. Note variability in the ∼500-bp band of T. dubius origin. (E) Southern blot hybridization showing both inheritance of parental and appearance of novel population-specific bands in population T. mirus 2602 (novel bands not present in any of the parental diploid accessions are indicated with asterisks).
Figure 4.—
Southern blot hybridization analysis of the IGS region. (A) Restriction enzyme maps of the major T. dubius, T. pratensis, and T. porrifolius rDNA units. The position of probe hybridization is indicated by a thin line. The sizes of hybridization fragments are indicated. 18S, 5.8S, and 26S are genes coding for rRNA molecules. (B) Southern blot hybridization of genomic DNAs showing variation in the parental IGS regions among populations of T. mirus. The DNAs were digested with the _Kpn_I restriction enzyme and hybridized with the 18S rDNA probe. The diagnostic fragments representing T. dubius (du) and T. porrifolius (po) families are indicated. T. porrifolius had a variable number of bands in the 8-kb region, indicating the presence of multiple gene families displaying sequence polymorphisms in the IGS region. Numbers in italics below the tracts indicate relative proportions of the _T. dubius_-specific band expressed as a percentage of total signal in the tract (also for C). (C) Southern blot hybridization of genomic DNAs showing variation in the IGS families among populations of T. miscellus. The digestion and hybridization conditions were the same as in B. The diagnostic fragments representing T. dubius (du) and T. pratensis (pr) families are indicated. (D) The presence of novel polymorphic sites in the IGS spacer of T. mirus, population 2602. Genomic DNAs from T. dubius, T. porrifolius, and T. mirus 2602-6 were digested with _Bst_YI/_Ssp_I and _Vsp_I methylation-insensitive restriction enzymes and the blots were hybridized to the 26S probe. Hybrid-specific bands not seen in any progenitor populations are indicated by asterisks.
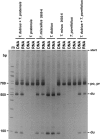
Figure 5.—
35S rRNA gene expression in diploid and allotetraploid species of Tragopogon. Primary RNA transcripts were analyzed in the ITS-1 region by RT-PCR using _Bst_NI restriction site polymorphisms. The digestion products of PCR reactions were separated on a 7% polyacrylamide gel and stained with ethidium bromide. The profiles after reverse transcription and subsequent cDNA amplification are indicated as “RNA”; the profiles obtained after DNA amplification are indicated as “DNA.” Control mixing experiments containing RNA and DNA samples are shown on the right and left margins. The sizes of the diagnostic bands were as follows: 90 bp (T. dubius), 500 bp (T. dubius), 680 bp (T. porrifolius), and 680 bp (T.pratensis).
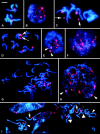
Figure 6.—
FISH to root-tip metaphase (A, C, D, G, and I), interphase (B, E, and F), and prophase (H) cells of Tragopogon using biotin-labeled pTa71 detected with avidin-Cy3 (red fluorescence) for 18-5.8-26S rDNA and counterstained with DAPI for DNA (blue fluorescence). (A and B) T. dubius at metaphase. (A) One pair of rDNA loci, which show variable levels of decondensation at interphase (B), giving strings and foci of signal. (C) T. pratensis has one pair of rDNA loci; the apparently remote signal on the metaphase chromosomes (arrows) indicates condensed rDNAs distal to secondary constrictions. (D and E) T. porrifolius. The metaphase preparation (D) shows two pairs of rDNA loci, one on chromosome A and one on chromosome D; each pair is potentially active, but often only the sites on chromosome D are decondensed, resulting in secondary constrictions (D, arrows). At interphase (E), the less active pair on chromosome A is regularly condensed and outside the nucleolus (arrowheads). (F and G) T. miscellus 2605, with two pairs of rDNA sites; both are decondensed at interphase (F), giving strings and foci of signal, and both have secondary constrictions at metaphase (G). (H and I) T. mirus 2602 has three pairs of rDNA loci. All are potentially active, but one pair is typically condensed and lacks secondary constrictions, identified at prophase (H, arrowheads) and metaphase (I, small arrowhead). In I, the arrows identify the rDNA loci of chromosome D of T. porrifolius. Assuming that rDNA condensation patterns are inherited in a manner that resembles those of the diploids, then the large arrowhead designates the _T. dubius_-derived locus and the small arrowhead the _T. porrifolius_-derived locus from chromosome A.
Similar articles
- Concerted evolution of rDNA in recently formed Tragopogon allotetraploids is typically associated with an inverse correlation between gene copy number and expression.
Matyásek R, Tate JA, Lim YK, Srubarová H, Koh J, Leitch AR, Soltis DE, Soltis PS, Kovarík A. Matyásek R, et al. Genetics. 2007 Aug;176(4):2509-19. doi: 10.1534/genetics.107.072751. Epub 2007 Jul 1. Genetics. 2007. PMID: 17603114 Free PMC article. - Similar patterns of rDNA evolution in synthetic and recently formed natural populations of Tragopogon (Asteraceae) allotetraploids.
Malinska H, Tate JA, Matyasek R, Leitch AR, Soltis DE, Soltis PS, Kovarik A. Malinska H, et al. BMC Evol Biol. 2010 Sep 22;10:291. doi: 10.1186/1471-2148-10-291. BMC Evol Biol. 2010. PMID: 20858289 Free PMC article. - Homeolog loss and expression changes in natural populations of the recently and repeatedly formed allotetraploid Tragopogon mirus (Asteraceae).
Koh J, Soltis PS, Soltis DE. Koh J, et al. BMC Genomics. 2010 Feb 8;11:97. doi: 10.1186/1471-2164-11-97. BMC Genomics. 2010. PMID: 20141639 Free PMC article. - On the origins of species: does evolution repeat itself in polyploid populations of independent origin?
Soltis DE, Buggs RJ, Barbazuk WB, Schnable PS, Soltis PS. Soltis DE, et al. Cold Spring Harb Symp Quant Biol. 2009;74:215-23. doi: 10.1101/sqb.2009.74.007. Epub 2009 Aug 17. Cold Spring Harb Symp Quant Biol. 2009. PMID: 19687140 Review. - Next-generation sequencing and genome evolution in allopolyploids.
Buggs RJ, Renny-Byfield S, Chester M, Jordon-Thaden IE, Viccini LF, Chamala S, Leitch AR, Schnable PS, Barbazuk WB, Soltis PS, Soltis DE. Buggs RJ, et al. Am J Bot. 2012 Feb;99(2):372-82. doi: 10.3732/ajb.1100395. Epub 2012 Jan 20. Am J Bot. 2012. PMID: 22268220 Review.
Cited by
- Polyploidy on islands - concerted evolution and gene loss amid chromosomal stasis.
Joshi P, Ansari H, Dickson R, Ellison NW, Skema C, Tate JA. Joshi P, et al. Ann Bot. 2023 Feb 7;131(1):33-44. doi: 10.1093/aob/mcac051. Ann Bot. 2023. PMID: 35390127 Free PMC article. - The fate of 35S rRNA genes in the allotetraploid grass Brachypodium hybridum.
Borowska-Zuchowska N, Kovarik A, Robaszkiewicz E, Tuna M, Tuna GS, Gordon S, Vogel JP, Hasterok R. Borowska-Zuchowska N, et al. Plant J. 2020 Aug;103(5):1810-1825. doi: 10.1111/tpj.14869. Epub 2020 Jul 3. Plant J. 2020. PMID: 32506573 Free PMC article. - Intragenomic rDNA variation - the product of concerted evolution, mutation, or something in between?
Wang W, Zhang X, Garcia S, Leitch AR, Kovařík A. Wang W, et al. Heredity (Edinb). 2023 Sep;131(3):179-188. doi: 10.1038/s41437-023-00634-5. Epub 2023 Jul 4. Heredity (Edinb). 2023. PMID: 37402824 Free PMC article. Review. - Angiosperms Are Unique among Land Plant Lineages in the Occurrence of Key Genes in the RNA-Directed DNA Methylation (RdDM) Pathway.
Ma L, Hatlen A, Kelly LJ, Becher H, Wang W, Kovarik A, Leitch IJ, Leitch AR. Ma L, et al. Genome Biol Evol. 2015 Sep 2;7(9):2648-62. doi: 10.1093/gbe/evv171. Genome Biol Evol. 2015. PMID: 26338185 Free PMC article. - A Robust Methodology for Assessing Differential Homeolog Contributions to the Transcriptomes of Allopolyploids.
Boatwright JL, McIntyre LM, Morse AM, Chen S, Yoo MJ, Koh J, Soltis PS, Soltis DE, Barbazuk WB. Boatwright JL, et al. Genetics. 2018 Nov;210(3):883-894. doi: 10.1534/genetics.118.301564. Epub 2018 Sep 13. Genetics. 2018. PMID: 30213855 Free PMC article.
References
- Abbott, R. J., and A. J. Lowe, 2004. Origins, establishment and evolution of two new polyploid species of Senecio in the British Isles. Biol. J. Linn. Soc. 82: 467–474.
- Ainouche, M. L., A. Baumel and A. Salmon, 2004. Spartina anglica Schreb.: a natural model system for analysing evolutionary changes that affect allopolyploid genomes. Biol. J. Linn. Soc. 82: 475–484.
- Álvarez, I., and J. F. Wendel, 2003. Ribosomal ITS sequences and plant phylogenetic inference. Mol. Phylogenet. Evol. 29: 417–434. - PubMed
- Bennett, R. I., and A. G. Smith, 1991. Use of a genomic clone for ribosomal RNA from Brassica oleracea in RFLP analysis of Brassica species. Plant. Mol. Biol. 16: 685–688. - PubMed
- Brehm, B. G., and M. Ownbey, 1965. Variation in chromatographic patterns in the Tragopogon dubius-pratensis-porrifolius complex (Compositae). Am. J. Bot. 52: 811–818.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources