Olig2 and Ngn2 function in opposition to modulate gene expression in motor neuron progenitor cells - PubMed (original) (raw)
Olig2 and Ngn2 function in opposition to modulate gene expression in motor neuron progenitor cells
Soo-Kyung Lee et al. Genes Dev. 2005.
Abstract
Spinal motor neurons and oligodendrocytes are generated sequentially from a common pool of progenitors termed pMN cells. Olig2 is a bHLH-class transcription factor in pMN cells, but it has remained unclear how its transcriptional activity is modulated to first produce motor neurons and then oligodendrocytes. Previous studies have shown that Olig2 primes pMN cells to become motor neurons by triggering the expression of Ngn2 and Lhx3. Here we show that Olig2 also antagonizes the premature expression of post-mitotic motor neuron genes in pMN cells. This blockade is counteracted by Ngn2, which accumulates heterogeneously in pMN cells, thereby releasing a subset of the progenitors to differentiate and activate expression of post-mitotic motor neuron genes. The antagonistic relationship between Ngn2 and Olig2 is mediated by protein interactions that squelch activity as well as competition for shared DNA-binding sites. Our data support a model in which the Olig2/Ngn2 ratio in progenitor cells serves as a gate for timing proper gene expression during the development of pMN cells: Olig2(high) maintains the pMN state, thereby holding cells in reserve for oligodendrocyte generation, whereas Ngn2(high) favors the conversion of pMN cells into post-mitotic motor neurons.
Figures
Figure 1.
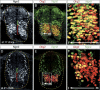
Immunohistochemical analysis of bHLH transcription factor expression in the developing chick neural tube. (A,B) Olig2 and Ngn2 are coexpressed in chick embryo pMN cells (red bracket, A) at HH stage 17. Ngn2 is also expressed in Olig2– pV2 cells (green bracket, A) and dorsally located post-mitotic cells. (C) Enlargement of the boxed area in B reveals that the relative levels of Ngn2 and Olig2 vary within individual pMN cells. This results in a mix of red (Olig2high) and yellow (Olig2+/Ngn2+) labeling. (D_–_E) At HH stage 21, Olig2+/Ngn2+ cells are located at the subventricular zone where motor neurons emerge. In contrast, Olig2+/Ngn2– cells are predominantly found medially in the proliferative ventricular zone. (F) Enlargement of the boxed area in E reveals reduced levels of Ngn2 in the Olig2+ pMN cells at HH stage 21. The medial (m) to lateral (l) orientation of the ventricular zone is shown in B, C, E, and F.
Figure 2.
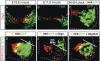
Olig2 attenuates the neurogenic activity of Ngn2. (A_–_D) Neuronal differentiation assays in transfected P19 cells cultured 3 d using anti-β tubulin neuronal marker antibody TuJ1 (red). (A) P19 cells transfected with GFP vector control lack TuJ1+ neurons. (B) Ngn2 triggers β-tubulin expression (red) in most transfected cells (green), and induces the formation of neurite processes (arrowheads). (C) Olig2 failed to drive neuronal differentiation in P19 cells. (D) Olig2 markedly reduced the ability of Ngn2 to induce β-tubulin expression and prevented the formation of neurite processes. (E) The neurogenic activity of Ngn2 and Olig2 was determined by calculating the percentage of transfected cells (GFP+) labeled by TuJ1 after 1 d in culture. (F_–_K) TuJ1 neuronal marker induction in chick embryo neural tubes 43 h post-electroporation. (F_–_H) Ngn2 induces ectopic and precocious TuJ1 neuronal marker expression. Many Ngn2+ cells coexpress TuJ1 (yellow) and TuJ1 labeling of neurons is detected across the medial (m) lateral (l) axis of the neural tube. The box in F corresponds to approximate regions shown in G and H. (I_–_K) Olig2 is less efficient than Ngn2 at driving neuronal differentiation. In contrast to Ngn2, Olig2+ cells rarely express TuJ1 and fewer TuJ1+ cells are detected across the m–l axis. The boxed area in I corresponds to images in J and K.
Figure 3.
Constitutive expression of Olig2 is detrimental to motor neuron generation. (A_–_F) Immunocytochemical analysis of motor neuron differentiation monitored using the Isl1 marker of these cells in HH stage 20 chick embryos electroporated with CMV–Olig2–ires–GFP. (A_–_C) In embryos with a high percentage of electroporated (GFP+) cells, Isl1 levels were markedly reduced in the endogenous motor neuron domain concomitant with development of ectopic motor neurons (ect. MNs) in the V2 domain. The box in B corresponds to the image in C.(D_–_F) Electroporated embryos with a mosaic distribution of Olig2 marked by more scattered GFP labeling exhibited a selective bias in the distribution of Olig2-expressing cells. Because of apparent sorting, motor neurons developed normally but were derived from cells lacking constitutive Olig2. Again, ectopic motor neurons were detected (ect. MNs). The box in E corresponds to image in F. (G) Motor neuron differentiation analysis (Hb9+ cells) in HH stage 22 chick embryos electroporated with the indicated LIM and bHLH constructs. Coelectroporation of Ngn2 with Isl1 and Lhx3 potentiates motor neuron generation, whereas Olig2 lacks this stimulatory effect. (H) Motor neuron induction assays in P19 cells. Olig2 expression alone or in combination with Ngn2 failed to trigger motor neuron differentiation. Unlike Ngn2, Olig2 is unable to synergize with Isl1 and Lhx3 to specify motor neurons.
Figure 4.
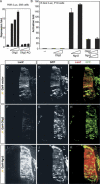
Olig2 represses gene expression in post-mitotic motor neurons. (A,B) Immunohistochemical analysis of embryonic day 10.5–11.5 (E10.5–E11.5) mouse ventral neural tube with antibodies against Olig2 (red) and the motor neuron marker Hb9 (green). pMN cells located laterally down-regulate Olig2 as they differentiate into Hb9+ motor neurons. In contrast, undifferentiated pMN cells within the ventricular zone maintain Olig2 expression. (C_–_F) GFP labeling driven from a reporter (Hb9::GFP) containing the motor neuron enhancer (M250 element, MNE) isolated from the Hb9 gene (Lee et al. 2004) in combination with Olig2 or Hb9/MNR2 expression in HH stage 24 chick embryos. MNR2 monoclonal antibody staining detects two Hb9-related proteins in chick, MNR2 in progenitors and young motor neurons, and to a lesser extent Hb9 in mature motor neurons (Tanabe et al. 1998). (C) Electroporation of Hb9::GFP results in GFP expression (green) in post-mitotic motor neurons but not Olig2+ (red) pMN cells. (D) Electroporation of the Hb9::GFP promoter construct labels only the lateral Hb9/MNR2+ cells (yellow). (E) Constitutive expression of Olig2 suppressed Hb9::GFP in post-mitotic motor neurons, but induced GFP expression in the V2 interneuron domain where ectopic motor neurons form (ect. MNs). (F) The Olig2-AQ DNA-binding mutant is less efficient at suppressing Hb9::GFP promoter activity in motor neurons and fails to induce ectopic expression of GFP in the V2 domain.
Figure 5.
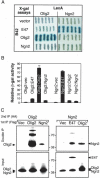
Olig2 interacts with bifunctional activator/suppressor E-boxes within Hb9. (A) Schematic representation of the M250 motor neuron enhancer element isolated from the Hb9 gene linked to the proximal region of the Hb9 promoter (ΔNhe). Two evolutionarily conserved E-boxes are located within the enhancer (M50 E-box and M100 E-box) (Lee et al. 2004). Wild-type and E-box mutants were electroporated into chick embryos, and the relative level of GFP in motor neurons (MN Exp.) and ectopic sites (Ect. Exp.) was monitored at HH stage 24. The table is representative of ∼10 embryos for each construct (+++ indicates strong expression; ++ indicates moderate expression; + indicates low-level expression). (B) The wild-type M250 enhancer in the Hb9::GFP reporter is inactive in the pMN cell domain marked by Olig2 expression (red). In contrast, mutation of the M50 E-box element within M250 results in the derepression of Hb9 expression in the progenitor cell domain. Likewise, mutation of the M100 E-box, and M50 and M100 E-boxes in combination, also led to ectopic expression in the pMN domain (A; data not shown). (C,D) Gel retardation assays using the M100A probe containing the M100 E-box from the Hb9 enhancer (Lee et al. 2004). (C) Olig2 binds to the M100A oligonucleotide, whereas the Olig2 DNA-binding mutant Olig2-AQ lacks this activity. The M100A DNA–Olig2 protein complex was supershifted by antibody against the Flag epitope tag, but not by control IgG antibody. Probes for the M50 E-box element exhibit the same pattern of Olig2 binding (data not shown). (D) The M100A DNA–Olig2 protein interaction was challenged by 20- or 100-fold molar excess of the unlabeled M100A probe (Self), E-box mutated M100A (E-mt), or unrelated oligo (Non-self). Olig2 interacts with DNA E-box elements in a sequence-specific manner. (E) A presumed Olig2/Olig2 homodimer and Olig2/E47 heterodimer bind to the E-box site within the M100A probe. Similar results were obtained with the E-box site within the M50 probe (data not shown). Flag-tagged Olig2 and HA-tagged E47 were translated in vitro separately and incubated with the M100A DNA. Supershift assays using antibodies against the HA or Flag epitope tags confirmed the protein composition in each complex. (F,G) Chromatin immunoprecipitation experiments in transfected P19 cells. (F) Flag-tagged Olig2 wt (wild type) is detected bound to the endogenous Hb9 promoter, whereas the DNA-binding mutant Olig2-AQ failed to interact with this DNA sequence. Western blot analysis detects similar levels of protein expression. (G) Flag-tagged Olig2 wt was transfected into P19 cells with two derivatives of the Hb9 promoter: one carrying the native motor neuron enhancer (M250 wt) and the other with mutated E-box elements (M250 Emt). Primers that were selective for the transfected Hb9 promoter construct were used as shown in the schematic. Olig2 failed to bind to the Hb9 promoter with mutated E-box elements.
Figure 6.
Olig2 self-associates and forms weak heterointeractions with E47 and Ngn2. (A,B) Protein interactions between bHLH factors assayed using the yeast two-hybrid assay. Olig2 protein interacts strongly with itself, and weakly with E47 and Ngn2. Ngn2 does not self-associate but forms high-affinity heterodimers with E47. (C) In vitro pull-down assays using Flag and HA epitope tags for immunoprecipitation. Proteins were co-translated with 35S-labeled methionine, then complexes were isolated with Flag antibody, dissociated, and HA-proteins were identified by immunopurification and electrophoresis. Olig2 associates strongly with itself. Olig2 and Ngn2 appear to interact with lower affinity.
Figure 7.
Olig2 and Ngn2 function antagonistically to control gene expression. (A) Olig2 strongly inhibited luciferase expression in 293 cells driven from the Hb9 motor neuron enhancer construct Hb9::Luc. The Olig2-AQ DNA-binding mutant was not able to repress luciferase expression. (B) Olig2 suppressed Ngn2-mediated _trans_-activation of an E-box reporter in 293 cells. (C_–_K) In vivo Gal4-based assays to measure corepressor function with a UAS-LacZ construct in HH stage 24 electroporated chick embryo neural tubes. CMV-GFP is included as an independent electroporation control in the assays. (C_–_E) Gal4 alone does not suppress LacZ expression. (F_–_H) The Gal4–Olig2 fusion strongly inhibits LacZ expression along the entire dorsal–ventral axis of the neural tube, but does not interfere with GFP. (I_–_K) Gal4–Ngn2 has the opposite effect of Olig2 and stimulates LacZ expression.
Figure 8.
Model of Olig2 and Ngn2 function in motor neuron specification. (A) A transcriptional cascade initiated by Shh and RA leads to the expression of Olig2 in pMN cells that serve as the precursors for motor neurons and oligodendrocytes. Olig2 occupies a key nodal point in the pathway, contributing to the regulation of homeodomain factors for motor neuron subtype specification (Mnr2, Lhx3, Isl1), and bHLH factors (Ngn2, NeuroM/D) for neurogenesis and synergistic cooperation with the homeodomain factors. Here we provide evidence that a key function of Olig2 is to maintain the progenitor cell state by repressing the expression of genes such as Hb9 found in post-mitotic motor neurons. (B) Olig2 and Ngn2 function antagonistically to either suppress or activate motor neuron gene expression, respectively. The fate decision of pMN cells to become post-mitotic motor neurons or remain in reserve for the later formation of oligodendrocytes is thought to depend on the relative levels of the repressor Olig2 to the activators Ngn2 and NeuroM/D. (C) Olig2 uses both DNA-dependent and DNA-independent mechanisms to suppress motor neuron gene expression in pMN cells. Protein–protein interactions between Olig2 and the activator proteins Ngn2 and E47 are likely to squelch Ngn2:E47 dimer formation and minimize their ability to bind and activate gene expression. In addition, the binding sites for Olig2 complexes and Ngn2 complexes overlap, suggesting they compete for their DNA targets.
Similar articles
- Coordinate regulation of motor neuron subtype identity and pan-neuronal properties by the bHLH repressor Olig2.
Novitch BG, Chen AI, Jessell TM. Novitch BG, et al. Neuron. 2001 Sep 13;31(5):773-89. doi: 10.1016/s0896-6273(01)00407-x. Neuron. 2001. PMID: 11567616 - Olig2-positive progenitors in the embryonic spinal cord give rise not only to motoneurons and oligodendrocytes, but also to a subset of astrocytes and ependymal cells.
Masahira N, Takebayashi H, Ono K, Watanabe K, Ding L, Furusho M, Ogawa Y, Nabeshima Y, Alvarez-Buylla A, Shimizu K, Ikenaka K. Masahira N, et al. Dev Biol. 2006 May 15;293(2):358-69. doi: 10.1016/j.ydbio.2006.02.029. Epub 2006 Apr 3. Dev Biol. 2006. PMID: 16581057 - Synchronization of neurogenesis and motor neuron specification by direct coupling of bHLH and homeodomain transcription factors.
Lee SK, Pfaff SL. Lee SK, et al. Neuron. 2003 Jun 5;38(5):731-45. doi: 10.1016/s0896-6273(03)00296-4. Neuron. 2003. PMID: 12797958 - An 'oligarchy' rules neural development.
Rowitch DH, Lu QR, Kessaris N, Richardson WD. Rowitch DH, et al. Trends Neurosci. 2002 Aug;25(8):417-22. doi: 10.1016/s0166-2236(02)02201-4. Trends Neurosci. 2002. PMID: 12127759 Review. - Olig gene function in CNS development and disease.
Ligon KL, Fancy SP, Franklin RJ, Rowitch DH. Ligon KL, et al. Glia. 2006 Jul;54(1):1-10. doi: 10.1002/glia.20273. Glia. 2006. PMID: 16652341 Review.
Cited by
- Specification of CNS glia from neural stem cells in the embryonic neuroepithelium.
Kessaris N, Pringle N, Richardson WD. Kessaris N, et al. Philos Trans R Soc Lond B Biol Sci. 2008 Jan 12;363(1489):71-85. doi: 10.1098/rstb.2006.2013. Philos Trans R Soc Lond B Biol Sci. 2008. PMID: 17282992 Free PMC article. Review. - Spatiotemporal development of spinal neuronal and glial populations in the Ts65Dn mouse model of Down syndrome.
Aziz NM, Klein JA, Brady MR, Olmos-Serrano JL, Gallo V, Haydar TF. Aziz NM, et al. J Neurodev Disord. 2019 Dec 16;11(1):35. doi: 10.1186/s11689-019-9294-9. J Neurodev Disord. 2019. PMID: 31839007 Free PMC article. - Human oligodendrocytes from embryonic stem cells: conserved SHH signaling networks and divergent FGF effects.
Hu BY, Du ZW, Li XJ, Ayala M, Zhang SC. Hu BY, et al. Development. 2009 May;136(9):1443-52. doi: 10.1242/dev.029447. Development. 2009. PMID: 19363151 Free PMC article. - A targeted neuroglial reporter line generated by homologous recombination in human embryonic stem cells.
Xue H, Wu S, Papadeas ST, Spusta S, Swistowska AM, MacArthur CC, Mattson MP, Maragakis NJ, Capecchi MR, Rao MS, Zeng X, Liu Y. Xue H, et al. Stem Cells. 2009 Aug;27(8):1836-46. doi: 10.1002/stem.129. Stem Cells. 2009. PMID: 19544414 Free PMC article. - Cre recombination-mediated cassette exchange for building versatile transgenic human embryonic stem cells lines.
Du ZW, Hu BY, Ayala M, Sauer B, Zhang SC. Du ZW, et al. Stem Cells. 2009 May;27(5):1032-41. doi: 10.1002/stem.38. Stem Cells. 2009. PMID: 19415769 Free PMC article.
References
- Appel B. and Eisen, J.S. 1998. Regulation of neuronal specification in the zebrafish spinal cord by Delta function. Development 125: 371–380. - PubMed
- Arber S., Han, B., Mendelsohn, M., Smith, M., Jessell, T.M., and Sockanathan, S. 1999. Requirement for the homeobox gene Hb9 in the consolidation of motor neuron identity. Neuron 23: 659–674. - PubMed
- Bertrand N., Castro, D.S., and Guillemot, F. 2002. Proneural genes and the specification of neural cell types. Nat. Rev. Neurosci. 3: 517–530. - PubMed
- Briscoe J. and Ericson, J. 2001. Specification of neuronal fates in the ventral neural tube. Curr. Opin. Neurobiol. 11: 43–49. - PubMed
- Briscoe J., Pierani, A., Jessell, T.M., and Ericson, J. 2000. A homeodomain protein code specifies progenitor cell identity and neuronal fate in the ventral neural tube. Cell 101: 435–445. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases