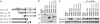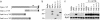Drosophila peptidoglycan recognition protein LC (PGRP-LC) acts as a signal-transducing innate immune receptor - PubMed (original) (raw)
Drosophila peptidoglycan recognition protein LC (PGRP-LC) acts as a signal-transducing innate immune receptor
Kwang-Min Choe et al. Proc Natl Acad Sci U S A. 2005.
Abstract
Drosophila peptidoglycan recognition protein LC (PGRP-LC), a transmembrane protein required for the response to bacterial infection, acts at the top of a cytoplasmic signaling cascade that requires the death-domain protein Imd and an IkappaB kinase to activate Relish, an NF-kappaB family member. It is not clear how binding of peptidoglycan to the extracellular domain of PGRP-LC activates intracellular signaling because its cytoplasmic domain has no homology to characterized proteins. Here, we demonstrate that PGRP-LC binds Imd and that its cytoplasmic domain is critical for its activity, suggesting that PGRP-LC acts as a signal-transducing receptor. The PGRP-LC cytoplasmic domain is also essential for the formation of dimers, and results suggest that dimerization may be required for receptor activation. The PGRP-LC cytoplasmic domain can mediate formation of heterodimers between different PGRP-LC isoforms, thereby potentially expanding the diversity of ligands that can be recognized by the receptor.
Figures
Fig. 1.
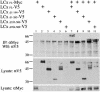
Domains of PGRP-LCx required for antimicrobial peptide gene induction. (a) PGRP-LC deletion constructs. PGRP-LC is a type II transmembrane protein with a previously uncharacterized N-terminal cytoplasmic domain, a transmembrane domain (TM) and a C-terminal PGRP domain. A V5 epitope was added in the C terminus of full-length and deletion constructs. (b) Protein expression in transiently transfected S2 cells was confirmed by Western blotting with anti-V5 antibody. (c) Induction of CecA1 was measured on Northern blots with RNA samples prepared from S2 cells expressing the PGRP-LCx deletion constructs. RNAs were purified from the cells after protein expression and, as shown in lanes 9–16, treatment with E. coli (102-fold dilution of OD600 = 1.5, 6 h of incubation). Rp49 was used as a loading control.
Fig. 2.
Domains of Imd required for antimicrobial peptide gene induction. (a) Imd deletion constructs. The C-terminal 80-aa death domain (DD) is shown. V5 tag was added in the C terminus of full-length and deletion constructs. (b) Protein expression in transiently transfected S2 cells was confirmed by Western blotting with anti-V5 antibody. (c) Induction of CecA1 was measured on Northern blots. Deletion of the N-terminal half, the C-terminal half including the death domain, or only the death domain all abolished CecA1 induction.
Fig. 3.
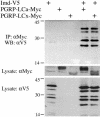
Physical interaction of PGRP-LC and Imd. Lysates were prepared from S2 cells transiently transfected with expression vectors for V5-tagged Imd and c-Myc-tagged PGRP-LCa or PGRP-LCx (20 μg each). Anti-c-Myc antibody was used to immunoprecipitate PGRP-LC, and anti-V5 antibody was used to detect Imd on Western blots.
Fig. 4.
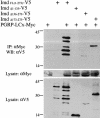
Identification of the domain of Imd that interacts with PGRP-LC. Lysates were prepared from cells transiently transfected with expression vectors for a V5-tagged Imd deletion construct and c-Myc-tagged PGRP-LCx (20 μg each). Anti-c-Myc antibody was used to immunoprecipitate the full-length PGRP-LCx, and anti-V5 antibody was used to detect Imd on Western blots. Deletion of the N-terminal half of Imd abolished the interaction, whereas deletion of the C-terminal half did not prevent interaction, suggesting that the N-terminal part is responsible for the interaction with PGRP-LC.
Fig. 5.
Homotypic and heterotypic interactions between PGRP-LCa and PGRP-LCx. Lysates were prepared from cells transiently transfected with expression vectors for the two PGRP-LC isoforms tagged with either V5 or c-Myc (20 μg each) and subjected to coimmunoprecipitation.
Fig. 6.
Mapping of the region of PGRP-LCx required for homotypic interactions. Lysates derived from cells transiently transfected with expression vectors for a V5-tagged deletion construct and the c-Myc-tagged full-length PGRP-LCx (20 μg each) were prepared and subjected to coimmunoprecipitation. Anti-c-Myc antibody was used to immunoprecipitate the full-length PGRP-LCx and anti-V5 antibody was used to detect the deletion constructs of PGRP-LCx by Western blotting. Note that deletion of the extracellular domain did not significantly affect the interaction, whereas deletion of the cytoplasmic domain abolished it (arrows mark IP products in lanes 7, 8, 10, and 11). Deletion of the distal half of the cytoplasmic domain did not prevent the interaction (lane 8), suggesting that the proximal half of the cytoplasmic domain is crucial for the interaction.
Similar articles
- PGRP-LC and PGRP-LE have essential yet distinct functions in the drosophila immune response to monomeric DAP-type peptidoglycan.
Kaneko T, Yano T, Aggarwal K, Lim JH, Ueda K, Oshima Y, Peach C, Erturk-Hasdemir D, Goldman WE, Oh BH, Kurata S, Silverman N. Kaneko T, et al. Nat Immunol. 2006 Jul;7(7):715-23. doi: 10.1038/ni1356. Epub 2006 Jun 11. Nat Immunol. 2006. PMID: 16767093 - Tissue- and ligand-specific sensing of gram-negative infection in drosophila by PGRP-LC isoforms and PGRP-LE.
Neyen C, Poidevin M, Roussel A, Lemaitre B. Neyen C, et al. J Immunol. 2012 Aug 15;189(4):1886-97. doi: 10.4049/jimmunol.1201022. Epub 2012 Jul 6. J Immunol. 2012. PMID: 22772451 - Peptidoglycan recognition protein (PGRP)-LE and PGRP-LC act synergistically in Drosophila immunity.
Takehana A, Yano T, Mita S, Kotani A, Oshima Y, Kurata S. Takehana A, et al. EMBO J. 2004 Nov 24;23(23):4690-700. doi: 10.1038/sj.emboj.7600466. Epub 2004 Nov 11. EMBO J. 2004. PMID: 15538387 Free PMC article. - Peptidoglycan recognition proteins: on and off switches for innate immunity.
Steiner H. Steiner H. Immunol Rev. 2004 Apr;198:83-96. doi: 10.1111/j.0105-2896.2004.0120.x. Immunol Rev. 2004. PMID: 15199956 Review. - [Intra- and extracellular recognition of pathogens and activation of innate immunity].
Kurata S. Kurata S. Yakugaku Zasshi. 2006 Dec;126(12):1213-8. doi: 10.1248/yakushi.126.1213. Yakugaku Zasshi. 2006. PMID: 17139146 Review. Japanese.
Cited by
- The role of the IAP E3 ubiquitin ligases in regulating pattern-recognition receptor signalling.
Vandenabeele P, Bertrand MJ. Vandenabeele P, et al. Nat Rev Immunol. 2012 Dec;12(12):833-44. doi: 10.1038/nri3325. Epub 2012 Nov 5. Nat Rev Immunol. 2012. PMID: 23124073 Review. - Drosophila as a Model for Human Viral Neuroinfections.
Benoit I, Di Curzio D, Civetta A, Douville RN. Benoit I, et al. Cells. 2022 Aug 29;11(17):2685. doi: 10.3390/cells11172685. Cells. 2022. PMID: 36078091 Free PMC article. Review. - Elevated CO2 suppresses specific Drosophila innate immune responses and resistance to bacterial infection.
Helenius IT, Krupinski T, Turnbull DW, Gruenbaum Y, Silverman N, Johnson EA, Sporn PH, Sznajder JI, Beitel GJ. Helenius IT, et al. Proc Natl Acad Sci U S A. 2009 Nov 3;106(44):18710-5. doi: 10.1073/pnas.0905925106. Epub 2009 Oct 21. Proc Natl Acad Sci U S A. 2009. PMID: 19846771 Free PMC article. - Toll and IMD pathways synergistically activate an innate immune response in Drosophila melanogaster.
Tanji T, Hu X, Weber AN, Ip YT. Tanji T, et al. Mol Cell Biol. 2007 Jun;27(12):4578-88. doi: 10.1128/MCB.01814-06. Epub 2007 Apr 16. Mol Cell Biol. 2007. PMID: 17438142 Free PMC article. - Continuous exposure to Plasmodium results in decreased susceptibility and transcriptomic divergence of the Anopheles gambiae immune system.
Aguilar R, Das S, Dong Y, Dimopoulos G. Aguilar R, et al. BMC Genomics. 2007 Dec 5;8:451. doi: 10.1186/1471-2164-8-451. BMC Genomics. 2007. PMID: 18053261 Free PMC article.
References
- Janeway, C. A., Jr., (1989) Cold Spring Harbor Symp. Quant. Biol. 54, 1–13. - PubMed
- Lemaitre, B., Nicolas, E., Michaut, L., Reichhart, J. M. & Hoffmann, J. A. (1996) Cell 86, 973–983. - PubMed
- Rutschmann, S., Kilinc, A. & Ferrandon, D. (2002) J. Immunol. 168, 1542–1546. - PubMed
- Levashina, E. A., Langley, E., Green, C., Gubb, D., Ashburner, M., Hoffmann, J. A. & Reichhart, J. M. (1999) Science 285, 1917–1919. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases