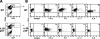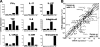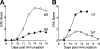IL-23 drives a pathogenic T cell population that induces autoimmune inflammation - PubMed (original) (raw)
IL-23 drives a pathogenic T cell population that induces autoimmune inflammation
Claire L Langrish et al. J Exp Med. 2005.
Abstract
Interleukin (IL)-23 is a heterodimeric cytokine composed of a unique p19 subunit, and a common p40 subunit shared with IL-12. IL-12 is important for the development of T helper (Th)1 cells that are essential for host defense and tumor suppression. In contrast, IL-23 does not promote the development of interferon-gamma-producing Th1 cells, but is one of the essential factors required for the expansion of a pathogenic CD4(+) T cell population, which is characterized by the production of IL-17, IL-17F, IL-6, and tumor necrosis factor. Gene expression analysis of IL-23-driven autoreactive T cells identified a unique expression pattern of proinflammatory cytokines and other novel factors, distinguishing them from IL-12-driven T cells. Using passive transfer studies, we confirm that these IL-23-dependent CD4(+) T cells are highly pathogenic and essential for the establishment of organ-specific inflammation associated with central nervous system autoimmunity.
Figures
Figure 1.
T cells and inflammatory macrophages enter the CNS of MOG-immunized IL-23p19 −/− mice. (A) Phenotypic analysis of CNS-infiltrating cells in the brains of WT and IL-23p19−/− mice at day 7 (before EAE onset) after immunization with MOG/CFA. (B) Intracellular cytokine staining of day 7 CNS-infiltrating cells from WT and IL-23p19−/− mice after 18 h of stimulation with 100 μg/ml MOG peptide, all plots are gated on live CD4+ T cells. Data are representative of five separate experiments.
Figure 2.
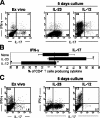
IL-23 promotes the expansion of IL-17 producing CD4 + (ThIL-17) cells. (A) Intracellular IL-17 and IFN-γ production by CD4+ DLN cells isolated from PLP-immunized WT SJL mice, either immediately after ex vivo isolation, or cultured with PLP139-151 peptide plus either rIL-12, rIL-23, or no added cytokine for 5 d. All plots are gated on live CD4+ T cells. (B) Mean percentage (±1 SD of error) of CD4+ T cells producing IFN-γ or IL-17 in response to 5 d of culture with IL-12 or IL-23. *, Student's unpaired t test, P < 0.02, averaged from eight separate experiments. (C) Intracellular IL-17 and IFN-γ production by CD4+ DLN cells isolated from MOG-immunized p40−/− C57BL/6 either immediately after ex vivo isolation or cultured for 5 d with rIL-12 or rIL-23. All intracellular staining samples were restimulated with PMA/ionomycin for 4 h before analysis. All plots are gated on live CD4+ T cells and are representative of three independent experiments.
Figure 3.
IL-23 drives a small population of naive T cells to produce IL-17 after in vitro activation. (A) Surface expression of CD62L on naive splenic DO11.10 × Rag−/− T cells, either analyzed immediately after isolation (ex vivo) or on day 7 after in vitro anti-CD3/anti-CD28 stimulation plus IL-12 or IL-23. (B) Intracellular cytokine staining of CD4+ DO11.10 × Rag−/− T cells on day 7 after in vitro anti-CD3/anti-CD28 stimulation plus IL-12 or IL-23. Cells were restimulated with PMA/ionomycin for 4 h before intracellular staining. (C) Naive DO11.10 × Rag−/− T cells were parked in BALB/c recipients and OVA/CFA immunized. DLN cells were restimulated, PMA/ionomycin and intracellular stained for IL-17 and IFN-γ or (D) surface stained for CD44 and CD62L. All plots are gated on CD4+ KJ1-26+ cells. Results are representative of two experiments.
Figure 4.
Gene expression analysis of IL-23 versus IL-12 driven T cells. (A) Quantitative analysis of mRNA expression levels of selected genes, comparing sorted CD4+ DLN cells isolated from PLP-immunized mice, either not cultured (ex vivo) or cultured with rIL-23 or rIL-12 for 10 d. (B) Quantitative analysis of mRNA expression levels of >200 selected genes, comparing total DLN cells isolated from PLP-immunized mice, cultured with either rIL-23 or rIL-12 for 10 d. All samples were activated with PMA/ionomycin for 4 h before RNA extraction and Taqman analysis. Results are normalized to a housekeeping gene, ubiquitin, and representative of two similar experiments.
Figure 5.
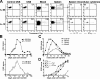
IL-17–producing ThIL-17 cells induce EAE. (A) PLP-primed DLN cells cultured with rIL-23 or rIL-12 for 10 d, labeled with CFSE (peak MFI = 4,654) and transferred (5 × 106 cells/mouse) into recipient WT SJL mice. Cells were tracked at day 3 after immunization, with further gating on splenic CFSE+ CD4+ cells to determine IFN-γ and IL-17 production by intracellular staining. (B) PLP-primed DLN cells cultured with either rIL-23 (squares) or rIL-12 (triangles) for 10 d, and transferred into WT SJL mice as either purified CD4+ cells (3 × 106 cells/mouse; n = 10/group; top), or as total DLN cells (7 × 106 cells/mouse; n = 3/group; bottom), with mean EAE scores recorded. (C) Mean EAE scores of WT recipient mice (n = 5/group) passively transferred with graded numbers of ThIL-17 cells: 1.2 × 106 ThIL-17 (closed squares), 3 × 105 ThIL-17 (closed triangles), 1.5 × 105 ThIL-17 (open circles), or 2.0 × 104 ThIL-17 (open diamonds). Results are summarized from two separate experiments. (D) Mean EAE scores of WT recipient mice (n = 5/group) passively transferred with either 5 × 105 ThIL-17 cells (open squares), 5 × 105 Th1 cells (open diamonds), or cotransferred with ThIL-17 plus Th1 cells (5 × 105 of each; closed triangles), or ThIL-17 plus sorted IFN-γ− cells (5 × 105 ThIL-17 plus 2 × 105 IFN-γ− cells; closed circles). (C–D) ThIL-17 cell numbers are calculated from intracellular cytokine staining analysis, with ∼3-5 × 106 total DLN cells passively transferred per recipient mouse.
Figure 6.
IL-17, not IFN-γ, contributes to EAE severity. (A) WT SJL mice actively immunized with PLP emulsified in CFA, and treated at day 7 with 200 μg anti–IL-17 (closed diamonds) or isotype control (open diamonds), n = 5/group. (B) WT SJL mice actively immunized with PLP emulsified in CFA (using suboptimal immunization strategy to give weaker disease), and treated at days 0 and 7 with 200 μg anti–IFN-γ (closed squares) or isotype control (open squares), n = 7/group. Plots are representative of three similar experiments.
Similar articles
- Pathophysiology of interleukin-23 in experimental autoimmune encephalomyelitis.
Touil T, Fitzgerald D, Zhang GX, Rostami AM, Gran B. Touil T, et al. Drug News Perspect. 2006 Mar;19(2):77-83. doi: 10.1358/dnp.2006.19.2.977443. Drug News Perspect. 2006. PMID: 16628262 Review. - IL-12- and IL-23-induced T helper cell subsets: birds of the same feather flock together.
Bettelli E, Kuchroo VK. Bettelli E, et al. J Exp Med. 2005 Jan 17;201(2):169-71. doi: 10.1084/jem.20042279. J Exp Med. 2005. PMID: 15657286 Free PMC article. - Interleukin-23 rather than interleukin-12 is the critical cytokine for autoimmune inflammation of the brain.
Cua DJ, Sherlock J, Chen Y, Murphy CA, Joyce B, Seymour B, Lucian L, To W, Kwan S, Churakova T, Zurawski S, Wiekowski M, Lira SA, Gorman D, Kastelein RA, Sedgwick JD. Cua DJ, et al. Nature. 2003 Feb 13;421(6924):744-8. doi: 10.1038/nature01355. Nature. 2003. PMID: 12610626 - Stepwise regulation of TH1 responses in autoimmunity: IL-12-related cytokines and their receptors.
Becker C, Wirtz S, Neurath MF. Becker C, et al. Inflamm Bowel Dis. 2005 Aug;11(8):755-64. doi: 10.1097/01.mib.0000172808.03877.4d. Inflamm Bowel Dis. 2005. PMID: 16043992 Review.
Cited by
- Ubiquitin-independent proteosomal degradation of myelin basic protein contributes to development of neurodegenerative autoimmunity.
Belogurov A Jr, Kuzina E, Kudriaeva A, Kononikhin A, Kovalchuk S, Surina Y, Smirnov I, Lomakin Y, Bacheva A, Stepanov A, Karpova Y, Lyupina Y, Kharybin O, Melamed D, Ponomarenko N, Sharova N, Nikolaev E, Gabibov A. Belogurov A Jr, et al. FASEB J. 2015 May;29(5):1901-13. doi: 10.1096/fj.14-259333. Epub 2015 Jan 29. FASEB J. 2015. PMID: 25634956 Free PMC article. - The myeloid receptor PILRβ mediates the balance of inflammatory responses through regulation of IL-27 production.
Tato CM, Joyce-Shaikh B, Banerjee A, Chen Y, Sathe M, Ewald SE, Liu MR, Gorman D, McClanahan TK, Phillips JH, Heyworth PG, Cua DJ. Tato CM, et al. PLoS One. 2012;7(3):e31680. doi: 10.1371/journal.pone.0031680. Epub 2012 Mar 27. PLoS One. 2012. PMID: 22479310 Free PMC article. - PRMT5 regulates epigenetic changes in suppressive Th1-like iTregs in response to IL-12 treatment.
Jadon N, Shanthalingam S, Tew GN, Minter LM. Jadon N, et al. Front Immunol. 2024 Jan 8;14:1292049. doi: 10.3389/fimmu.2023.1292049. eCollection 2023. Front Immunol. 2024. PMID: 38259494 Free PMC article. - IL-23 signaling is not an important driver of liver inflammation and fibrosis in murine non-alcoholic steatohepatitis models.
Heredia JE, Sorenson C, Flanagan S, Nunez V, Jones C, Martzall A, Leong L, Martinez AP, Scherl A, Brightbill HD, Ghilardi N, Ding N. Heredia JE, et al. PLoS One. 2022 Sep 15;17(9):e0274582. doi: 10.1371/journal.pone.0274582. eCollection 2022. PLoS One. 2022. PMID: 36107926 Free PMC article. - TH17 cell: a double-edged sword in the development of inflammatory bowel disease.
Wen Y, Wang H, Tian D, Wang G. Wen Y, et al. Therap Adv Gastroenterol. 2024 Feb 21;17:17562848241230896. doi: 10.1177/17562848241230896. eCollection 2024. Therap Adv Gastroenterol. 2024. PMID: 38390028 Free PMC article. Review.
References
- Cua, D.J., J. Sherlock, Y. Chen, C.A. Murphy, B. Joyce, B. Seymour, L. Lucian, W. To, S. Kwan, T. Churakova, et al. 2003. Interleukin-23 rather than interleukin-12 is the critical cytokine for autoimmune inflammation of the brain. Nature. 421:744–748. - PubMed
- Oppmann, B., R. Lesley, B. Blom, J.C. Timans, Y. Xu, B. Hunte, F. Vega, N. Yu, J. Wang, K. Singh, et al. 2000. Novel p19 protein engages IL-12p40 to form a cytokine, IL-23, with biological activities similar as well as distinct from IL-12. Immunity. 13:715–725. - PubMed
- O'Garra, A., and N. Arai. 2000. The molecular basis of T helper 1 and T helper 2 cell differentiation. Trends Cell Biol. 10:542–550. - PubMed
- Shtrichman, R., and C.E. Sanuel. 2001. The role of gamma interferon in antimicrobial immunity. Curr. Opin. Microbiol. 4:251–259. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials