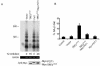POT1 and TRF2 cooperate to maintain telomeric integrity - PubMed (original) (raw)
POT1 and TRF2 cooperate to maintain telomeric integrity
Qin Yang et al. Mol Cell Biol. 2005 Feb.
Abstract
Mammalian telomeric DNA contains duplex TTAGGG repeats and single-stranded overhangs. POT1 (protection of telomeres 1) is a telomere-specific single-stranded DNA-binding protein, highly conserved in eukaryotes. The biological function of human POT1 is not well understood. In the present study, we demonstrate that POT1 plays a key role in telomeric end protection. The reduction of POT1 by RNA interference led to the loss of telomeric single-stranded overhangs and induced apoptosis, chromosomal instability, and senescence in cells. POT1 and TRF2 interacted with each other to form a complex with telomeric DNA. A dominant negative TRF2, TRF2(DeltaBDeltaM), bound to POT1 and prevented it from binding to telomeres. POT1 overexpression protected against TRF2(DeltaBDeltaM)-induced loss of telomeric single-stranded overhangs, chromosomal instability, and senescence. These results demonstrate that POT1 and TRF2 share in part in the same pathway for telomere capping and suggest that POT1 binds to the telomeric single-stranded DNA in the D-loop and cooperates with TRF2 in t-loop maintenance.
Figures
FIG. 1.
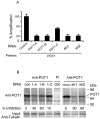
Reduction of POT1 expression by POT1-RNAi. (A) IMR90 cells were transduced with three POT1-siRNAs (POT1-A, -B, and -C) or two lentivirus shRNAs (sh1 and sh2); a random sequence siRNA was used as a control. After 48 h, RNA was prepared, and real-time reverse transcription-PCR was performed with the POT1 primers and probe. (B) POT1 expression was decreased by expression of POT1-RNAi. After expression for 2 days, IMR90 cell extracts were subjected to immunoprecipitation with the anti-POT1 antibody, followed by Western blotting with the anti-POT1 antibody. PI, preimmune serum. Fifty micrograms of cell extract was loaded as input and analyzed by anti-γ-tubulin antibody.
FIG. 2.
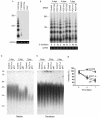
The expression of POT1-RNAi caused the loss of telomeric overhang signals. (A) T-OLAs were performed with oligonucleotides complementary to the G-rich strand [CCCTAA]4, the C-rich strand [TTAGGG]4, or the mismatch sequence [CCCTTA]4. (B) T-OLA analysis was used on DNA derived from IMR90 cells expressing POT1-siRNA and controls at 3, 5, and 7 days after transfection. Quantitative PCR was performed using primers specific for genomic GAPDH for equal amounts of genomic DNA in each sample (bottom). (C) The in-gel hybridization assay was performed with a native gel and probed with [CCCTTA]4. DNA was derived from IMR90 cells expressing POT1-RNAi. (D) The DNA was denatured in the gels and rehybridized with the same probes. (E) Quantitative data of the loss of telomeric overhangs were derived from three independent experiments similar to those shown in panel C, and the average value (error bars represent standard deviations) was plotted.
FIG. 3.
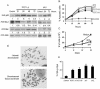
POT1-RNAi induced the DNA damage pathway, apoptosis, chromosomal abnormalities, and the senescent phenotype. (A) The time course of Western blotting analysis in MCF-7 cells expressing POT1-siRNA (POT1-A) or lentivirus POT1-shRNA (sh1). A random sequence siRNA was used as a control. (B) TUNEL assay of MCF7 cells after expression with POT1-siRNAs or shRNAs. A random sequence siRNA was used as a control. (C) Growth curve of IMR90 cells expressing lentivirus POT1-shRNAs and vector control cells. (D) The induction of chromosomal abnormalities. IMR90 cells were transduced with indicated siRNA or shRNA for 7 days, and the chromosome was prepared and analyzed as described in Materials and Methods. Open arrow, chromosome break; solid arrow, dicentric chromosome. (E) POT1-RNAi induced the senescent phenotype. IMR90 cells were stained for β-Gal activity at pH 6.0. Graphs show the effect of expression of POT1-siRNAs and shRNAs.
FIG. 4.
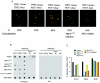
POT1 interacted with TRF2. (A) Endogenous TRF2 bound to POT1 in IMR90 fibroblasts by coimmunoprecipitation. Nuclear extracts (0.5 mg) were subjected to immunoprecipitation with the anti-POT1 antibody, followed by Western blotting with anti-TRF2, anti-RAD50, anti-NBS1, or anti-TRF1 antibodies. Preimmune serum (PI) was used as a control. Fifty micrograms of nuclear extract was loaded as input. (B) TRF2 and TRF2ΔBΔM bound to POT1 in 293 cells. Myc-TRF2 and TRF2ΔBΔM were transfected into 293 cells. Myc-TRF2 and TRF2ΔBΔM were coimmunoprecipitated with an anti-POT1 antibody from 0.5 mg of nuclear extract. Immunoprecipitated proteins were visualized by Western blotting analyses with antibodies against the Myc peptide. Myc-RPA1 was used as a control. (C) Myc-TRF2 and Myc-TRF2ΔBΔM brought down POT1-V5 (top) coexpressing POT1-V5 and Myc-TRF2 or Myc-TRF2ΔBΔM in 293 cells. TRF1 brought down POT1-V5 in 293 cells (bottom) expressing the POT1-V5 fusion protein. (D) POT1 interacted with TRF2 and TRF2ΔBΔM in vitro. Two micrograms of GST-TRF2 and TRF2ΔBΔM fusion proteins was incubated with 5 μl in vitro-translated POT1 protein labeled with [35S]methionine, and the proteins were brought down by GST beads. A 20% input of POT1 protein was included in this lane. GST-TRF2 and TRF2ΔBΔM protein inputs were verified by Coomassie blue staining. (E) POT1 and TRF2 formed a complex with telomeric single-stranded overhang DNA (TTAGGG probe, 200 pM). EMSA assay with increasing amounts of TRF2 (lanes 3 to 6, threefold steps up to 80 pM) with 20 pM POT1. Unlabeled [TTAGGG]4, 10-fold excess. (F) POT1 and TRF2 bound on [TTAGGG]4 oligomers. The biotinylated [TTAGGG]4 (10 nM) was attached to streptavidin beads. POT1 (40 pM) and/or TRF2 (40 pM) was added and incubated with the [TTAGGG]4-coated beads. POT1 and TRF2 associated with DNA were detected by immunoblotting.
FIG. 5.
(A) POT1 colocalized with endogenous TRF1 and TRF2 in IMR90 fibroblasts by immunostaining with anti-TRF1, anti-TRF2, and anti-POT1 antibodies. POT1 reduced the colocalization with TRF1 2 days after expressing TRF2ΔBΔM or TRF2-siRNA. (B) POT1 associated with telomeric DNA. ChIP of HT1080 cells expressing pLPC-Myc, Myc-POT1, Myc-TRF2ΔBΔM, or POT1-A-siRNA with indicated antibodies or preimmune serum (PI) of POT1 antibody for 2 days after infection. Duplicate dot blots were probed for telomeric or Alu repeats. (C) The quantification of the data in panel B representing the percentage of TTAGGG DNA recovered in each sample. Averaged signals obtained with total DNA samples were used as 100% value for the quantification.
FIG. 6.
(A) The expression of POT1 decreased the loss of telomeric overhang induced by TRF2ΔBΔM. (Top) The T-OLA was used on genomic DNA derived from the IMR90 cells expressing the indicated constructs for 5 days after infection. Lane 1, no genomic DNA. (Middle) Quantitative PCR was performed, using primers specific for genomic GAPDH for equal amounts of genomic DNA in each sample. (Bottom) Myc-POT1- and Myc-TRF2ΔBΔM-expressing levels were determined by immunoblotting. (B) POT1 blocked the senescent phenotype induced by TRF2ΔBΔM. IMR90 cells were stained for β-Gal activity at pH 6.0. Graphs showed the effect of infection of Myc-TRF2ΔBΔM (P < 0.01), Myc-TRF2ΔBΔM plus Myc-POT1 (_P_ > 0.5), Myc-POT1 (P > 0.5), empty vector (P > 0.5), and the untreated control for 16 days. The data show the results of three experiments; the error bars represent standard deviations. P values were determined by Student's t test representing each indicated group and untreated control group. The P value between Myc-TRF2ΔBΔM and Myc-TRF2ΔBΔM plus Myc-POT1 was <0.05.
Similar articles
- The G-quadruplex ligand telomestatin inhibits POT1 binding to telomeric sequences in vitro and induces GFP-POT1 dissociation from telomeres in human cells.
Gomez D, O'Donohue MF, Wenner T, Douarre C, Macadré J, Koebel P, Giraud-Panis MJ, Kaplan H, Kolkes A, Shin-ya K, Riou JF. Gomez D, et al. Cancer Res. 2006 Jul 15;66(14):6908-12. doi: 10.1158/0008-5472.CAN-06-1581. Cancer Res. 2006. PMID: 16849533 - A RAP1/TRF2 complex inhibits nonhomologous end-joining at human telomeric DNA ends.
Bae NS, Baumann P. Bae NS, et al. Mol Cell. 2007 May 11;26(3):323-34. doi: 10.1016/j.molcel.2007.03.023. Mol Cell. 2007. PMID: 17499040 - TRF2 promotes dynamic and stepwise looping of POT1 bound telomeric overhang.
Paul T, Liou W, Cai X, Opresko PL, Myong S. Paul T, et al. Nucleic Acids Res. 2021 Dec 2;49(21):12377-12393. doi: 10.1093/nar/gkab1123. Nucleic Acids Res. 2021. PMID: 34850123 Free PMC article. - Shelterin proteins and cancer.
Patel TN, Vasan R, Gupta D, Patel J, Trivedi M. Patel TN, et al. Asian Pac J Cancer Prev. 2015;16(8):3085-90. doi: 10.7314/apjcp.2015.16.8.3085. Asian Pac J Cancer Prev. 2015. PMID: 25921101 Review. - Telomere biology: a new player in the end zone.
Colgin L, Reddel R. Colgin L, et al. Curr Biol. 2004 Oct 26;14(20):R901-2. doi: 10.1016/j.cub.2004.09.075. Curr Biol. 2004. PMID: 15498484 Review.
Cited by
- MiR-185 targets POT1 to induce telomere dysfunction and cellular senescence.
Li T, Luo Z, Lin S, Li C, Dai S, Wang H, Huang J, Ma W, Songyang Z, Huang Y. Li T, et al. Aging (Albany NY). 2020 Jul 18;12(14):14791-14807. doi: 10.18632/aging.103541. Epub 2020 Jul 18. Aging (Albany NY). 2020. PMID: 32687062 Free PMC article. - SmedOB1 is Required for Planarian Homeostasis and Regeneration.
Yin S, Huang Y, Zhangfang Y, Zhong X, Li P, Huang J, Liu D, Songyang Z. Yin S, et al. Sci Rep. 2016 Sep 22;6:34013. doi: 10.1038/srep34013. Sci Rep. 2016. PMID: 27654173 Free PMC article. - The crosstalk between telomeres and DNA repair mechanisms: an overview to mammalian somatic cells, germ cells, and preimplantation embryos.
Tire B, Talibova G, Ozturk S. Tire B, et al. J Assist Reprod Genet. 2024 Feb;41(2):277-291. doi: 10.1007/s10815-023-03008-2. Epub 2024 Jan 2. J Assist Reprod Genet. 2024. PMID: 38165506 Review. - Distinct requirements for Pot1 in limiting telomere length and maintaining chromosome stability.
Bunch JT, Bae NS, Leonardi J, Baumann P. Bunch JT, et al. Mol Cell Biol. 2005 Jul;25(13):5567-78. doi: 10.1128/MCB.25.13.5567-5578.2005. Mol Cell Biol. 2005. PMID: 15964812 Free PMC article. - DNA damage response at functional and dysfunctional telomeres.
Longhese MP. Longhese MP. Genes Dev. 2008 Jan 15;22(2):125-40. doi: 10.1101/gad.1626908. Genes Dev. 2008. PMID: 18198332 Free PMC article. Review.
References
- Baumann, P., and T. R. Cech. 2001. Pot1, the putative telomere end-binding protein in fission yeast and humans. Science 292:1171-1175. - PubMed
- Blackburn, E. H. 2000. Telomere states and cell fates. Nature 408:53-56. - PubMed
- Blackburn, E. H. 2001. Switching and signaling at the telomere. Cell 106:661-673. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous