Cellular transformation by the MSP58 oncogene is inhibited by its physical interaction with the PTEN tumor suppressor - PubMed (original) (raw)
Cellular transformation by the MSP58 oncogene is inhibited by its physical interaction with the PTEN tumor suppressor
Koichi Okumura et al. Proc Natl Acad Sci U S A. 2005.
Abstract
The PTEN (phosphatase and tensin homologue) tumor suppressor protein contains a single catalytic domain with both lipid and protein phosphatase activities. The remaining C-terminal half of the PTEN protein plays a role in its stability and is mutated in many clinical cancer samples. Here, we report that the PTEN C-terminal domain physically interacts with the forkhead-associated domain of the oncogenic MSP58 protein and that this interaction requires PTEN Thr-366. We further show that while MSP58 transforms Pten-/- mouse embryo fibroblasts (MEFs), concurrent introduction of wild-type PTEN causes a dramatic reduction in the number of MSP58-induced transformed foci. This PTEN-mediated inhibition of cellular transformation requires physical interaction as evidenced by the failure of PTEN(T366A) point mutation (residing within the MSP58 interaction domain) to suppress MSP-58-driven transformation. These observations, together with the capacity of catalytically inactive PTEN mutant (G129R) to suppress MSP58 oncogenicity, support the view that the C-terminal region of PTEN directly provides a previously uncharacterized biological function in its ability to regulate cellular transformation.
Figures
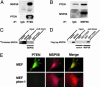
Fig. 1.
PTEN interacts with MSP58. (A) 293T cell lysates were precipitated with anti-PTEN or control IgG antibody followed by immunoblotting for coprecipitated MSP58. (B) Similarly, lysates were precipitated with anti-MSP58 followed by immunoblotting for coprecipitated PTEN. Efficiency of immunoprecipitation was assessed by reprobing each blot with antibodies used for precipitation. (C) 35S-labeled MSP58 protein, produced by TNT T7-coupled reticulocyte lysate, was applied to GST or GST-PTEN prebound to glutathione Sepharose (GST) beads for pull-down assays. (D) Lysates of 293T cells, which were transfected with empty vector or Flag-MSP58 plasmid, were applied to GST beads for pull-down assays. The pull-down product was detected by immunoblotting with anti-Flag tag horseradish peroxidase (HRP)-conjugated antibody. (E) Immunostaining for PTEN and MSP58 MEFs wild-type or null for PTEN.
Fig. 2.
MSP58 binds to the C terminus of PTEN through the MSP58 FHA domain. Lysates of 293T cells, transfected with various combinations of GFP-tagged (PTEN, full-length, amino acids 1–403; NPTEN, amino acids 1–202; PTENC, amino acids 186–403) and Flag-tagged (MSP58, full-length, amino acids 1–462; N-MSP58, amino acids 1–290; MSP58-C, amino acids 290–462) expression plasmids, were used in a coimmunoprecipitation (IP) assay. (A) Lysates of 293T cells were subjected to coimmunoprecipitation with anti-GFP antibody-conjugated agarose beads. Coimmunoprecipitated products were detected by immunoblotting with anti-Flag antibody. For confirmation of IP efficiency, the membrane was stripped and reprobed with anti-GFP antibody. (B) Lysates were incubated with anti-Flag antibody-conjugated agarose. Coimmunoprecipitated products were detected by immunoblotting with anti-GFP horseradish peroxidase (HRP)-conjugated antibody. For confirmation of IP efficiency, the membrane was stripped and reprobed with anti-Flag HRP-conjugated antibody.
Fig. 3.
Thr-366 of PTEN is critical for binding to MSP58. (A) Schematic diagram of phosphorylation sites in the PTEN C-terminal region and the FHA domains of various proteins displaying similarity to PTEN. (B) Lysates of 293T cells, cotransfected with MSP58 and various PTEN mutant expression plasmids, were used in a coimmunoprecipitation (IP) assay. Lysates were incubated with anti-Flag antibody conjugated agarose. Coimmunoprecipitated products were detected by immunoblotting with anti-GFP horseradish peroxidase-conjugated antibody. (Lower) Immunoblotting of GFP PTEN mutants for expression control.
Fig. 4.
PTEN interaction inhibits focus formation by MSP58. _Pten_–/– MEFs were infected with viruses expressing the indicated proteins. The cells were incubated for 40 days and then fixed and stained with crystal violet.
Comment in
- Phosphatase and tensin homologue growth suppression without phosphatase.
Stokoe D, Costello JF. Stokoe D, et al. Proc Natl Acad Sci U S A. 2005 Feb 22;102(8):2677-8. doi: 10.1073/pnas.0500089102. Epub 2005 Feb 14. Proc Natl Acad Sci U S A. 2005. PMID: 15710873 Free PMC article. No abstract available.
Similar articles
- Phosphatase and tensin homologue growth suppression without phosphatase.
Stokoe D, Costello JF. Stokoe D, et al. Proc Natl Acad Sci U S A. 2005 Feb 22;102(8):2677-8. doi: 10.1073/pnas.0500089102. Epub 2005 Feb 14. Proc Natl Acad Sci U S A. 2005. PMID: 15710873 Free PMC article. No abstract available. - PTEN: a novel anti-oncogenic function independent of phosphatase activity.
Okumura K, Zhao M, DePinho RA, Furnari FB, Cavenee WK. Okumura K, et al. Cell Cycle. 2005 Apr;4(4):540-2. doi: 10.4161/cc.4.4.1614. Epub 2005 Apr 21. Cell Cycle. 2005. PMID: 15753657 Review. - An essential role for protein synthesis in oncogenic cellular transformation.
Bader AG, Vogt PK. Bader AG, et al. Oncogene. 2004 Apr 19;23(18):3145-50. doi: 10.1038/sj.onc.1207550. Oncogene. 2004. PMID: 15094764 Review. - Pten signaling in gliomas.
Knobbe CB, Merlo A, Reifenberger G. Knobbe CB, et al. Neuro Oncol. 2002 Jul;4(3):196-211. Neuro Oncol. 2002. PMID: 12084351 Free PMC article. Review. - Regulation of cell migration by the C2 domain of the tumor suppressor PTEN.
Raftopoulou M, Etienne-Manneville S, Self A, Nicholls S, Hall A. Raftopoulou M, et al. Science. 2004 Feb 20;303(5661):1179-81. doi: 10.1126/science.1092089. Science. 2004. PMID: 14976311
Cited by
- Identification and characterization of nuclear and nucleolar localization signals in 58-kDa microspherule protein (MSP58).
Yang CP, Chiang CW, Chen CH, Lee YC, Wu MH, Tsou YH, Yang YS, Chang WC, Lin DY. Yang CP, et al. J Biomed Sci. 2015 May 16;22(1):33. doi: 10.1186/s12929-015-0136-0. J Biomed Sci. 2015. PMID: 25981436 Free PMC article. - Nuclear localization of PTEN by a Ran-dependent mechanism enhances apoptosis: Involvement of an N-terminal nuclear localization domain and multiple nuclear exclusion motifs.
Gil A, Andrés-Pons A, Fernández E, Valiente M, Torres J, Cervera J, Pulido R. Gil A, et al. Mol Biol Cell. 2006 Sep;17(9):4002-13. doi: 10.1091/mbc.e06-05-0380. Epub 2006 Jun 28. Mol Biol Cell. 2006. PMID: 16807353 Free PMC article. - Identification of novel PTEN-binding partners: PTEN interaction with fatty acid binding protein FABP4.
Gorbenko O, Panayotou G, Zhyvoloup A, Volkova D, Gout I, Filonenko V. Gorbenko O, et al. Mol Cell Biochem. 2010 Apr;337(1-2):299-305. doi: 10.1007/s11010-009-0312-1. Mol Cell Biochem. 2010. PMID: 19911253 - Synthetic lethal approaches to target cancers with loss of PTEN function.
Ertay A, Ewing RM, Wang Y. Ertay A, et al. Genes Dis. 2023 Nov;10(6):2511-2527. doi: 10.1016/j.gendis.2022.12.015. Genes Dis. 2023. PMID: 37533462 Free PMC article.
References
- Maehama, T., Taylor, G. S. & Dixon, J. E. (2001) Annu. Rev. Biochem. 70, 247–279. - PubMed
- Eng, C. (2003) Hum. Mutat. 22, 183–198. - PubMed
- Larsen, M., Tremblay, M. L. & Yamada, K. M. (2003) Nat. Rev. Mol. Cell Biol. 4, 700–711. - PubMed
- Lee, J. O., Yang, H., Georgescu, M. M., Di Cristofano, A., Maehama, T., Shi, Y., Dixon, J. E., Pandolfi, P. & Pavletich, N. P. (1999) Cell 99, 323–334. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials