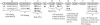Mutational analysis of the DEAD-box RNA helicase eIF4AII characterizes its interaction with transformation suppressor Pdcd4 and eIF4GI - PubMed (original) (raw)
Mutational analysis of the DEAD-box RNA helicase eIF4AII characterizes its interaction with transformation suppressor Pdcd4 and eIF4GI
Halina Zakowicz et al. RNA. 2005 Mar.
Abstract
Eukaryotic initiation factor (eIF) 4A unwinds secondary and tertiary structures in the 5'-untranslated region of mRNA, permitting translation initiation. Programmed cell death 4 (Pdcd4) is a novel transformation suppressor and eIF4A-binding partner that inhibits eIF4A helicase activity and translation. To elucidate the regions of eIF4A that are functionally significant in binding to Pdcd4, we generated point mutations of eIF4A. Two-hybrid analysis revealed that five eIF4A mutants completely lost binding to Pdcd4 while four eIF4A mutants retained wild-type levels of binding. The residues that, when mutated, inactivated Pdcd4 binding specified ATP binding, ATP hydrolysis, or RNA binding. With the exception of the Q-motif mutant eIF4AP56L, the eIF4A mutants inactivated for Pdcd4 binding were inactivated for binding to eIF4G (GM, GC, or both) and for enhancing translation. Several eIF4A mutants showing wild-type level binding to Pdcd4 were also inactivated for binding to eIF4G and for enhancing translation. Thus, significant dissociation of eIF4A's Pdcd4- and eIF4G-binding regions appears to occur. Because three of the four eIF4A mutants that retained Pdcd4 binding also suppressed translation activity in a dominant-negative manner, the structure that defines the Pdcd4-binding domain of eIF4A may be necessary but is insufficient for translation. A structural homology model of eIF4A shows regions important for binding to Pdcd4 and/or eIF4G lying on the perimeters of the hinge area of eIF4A. A competition experiment revealed that Pdcd4 competes with C-terminal eIF4G for binding to eIF4A. In summary, the Pdcd4-binding domains on eIF4A impact both binding to eIF4G and translation initiation in cells.
Figures
FIGURE 1.
The 15 point mutations of eIF4A, shown in bold letters where an amino acid has been changed. At the top is shown wild-type eIF4A with its 10 highly conserved domains.
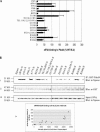
FIGURE 2.
Mutational analysis of eIF4A/Pdcd4 interaction. (A) Mammalian two-hybrid analysis of wild-type and mutant eIF4A interaction with Pdcd4. Two-hundred-fifty nanograms of wild-type/mutant pCMV-AD-eIF4A and 250 ng of pCMV-BD-Pdcd4 were transfected into RT101 cells, alongside 25 ng of Gal4-luciferase reporter gene and 5 ng of thymidine kinase-Renilla luciferase gene. Values (relative light units, or RLUs) are corrected for Renilla signal and provided as percentages of wild-type (WT) eIF4A interaction with Pdcd4. These experiments were repeated three times in sextuplicate, and representative data are shown. The results are expressed as the mean ± standard deviation. (B) Pull-down verification of eIF4A/mutant-xpress interaction levels with Pdcd4. (i) One-hundred micrograms of GST-Pdcd4 were added to 100 μg of RT101 cell lysates, which were then blotted with xpress antibody. (ii) Blots were stripped and probed with GST antibody to confirm equal affinity of glutathione-Sepharose beads for GST-Pdcd4. (iii) One-hundred microgram amounts of RT101 cell lysates were blotted with xpress antibody to show equal expression and loading of xpress-labeled protein. (iv) Densitometry readings were taken of each band in panel Bi, and the values were plotted against the RLU values in A.
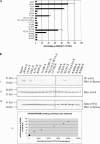
FIGURE 3.
Mutational analysis of eIF4A/eIF4GM(497–974) interaction. (A) Mammalian two-hybrid analysis of wild-type (WT) and mutant eIF4A interaction with eIF4G(497–974). Here 50 ng of wild-type/mutant pCMV-AD-eIF4A and 400 ng of pCMV-BD-eIF4G(497–974) were transfected into RT101 cells, alongside 25 ng of Gal4-luciferase reporter gene and 5 ng of thymidine kinase-Renilla luciferase gene. Values (relative light units, or RLUs) are corrected for Renilla signal and provided as percentages of wild-type eIF4A interaction with eIF4G(497–974). These experiments were repeated three times in sextuplicate, and representative data are shown. The results are expressed as the mean ± standard deviation. (B) Immunoprecipitation of eIF4A/mutant-xpress with eIF4G(497–974). (i) Here 2 μg of HA antibody was added to 100 μg of RT101 cell lysates, which were then blotted with xpress antibody. (ii) Blots were stripped and probed with HA antibody to confirm equal G-Sepharose bead affinity for HA antibody. (iii) One-hundred microgram amounts of RT101 cell lysates were blotted with xpress antibody to show equal expression and loading of xpress-labeled protein. (iv) Densitometry readings were taken of each band in panel Bi, and the values were plotted against the RLU values in A.
FIGURE 4.
(A) Mammalian two-hybrid analysis of wild-type (WT) and mutant eIF4A interaction with eIF4G(924–1444). Fifty nanograms of wild-type/mutant pCMV-AD-eIF4A and 400 ng of pCMV-BD-eIF4G(924–1444) were transfected into RT101 cells, alongside 25 ng of Gal4-luciferase reporter gene and 5 ng of thymidine kinase-Renilla luciferase gene. Values (relative light units, or RLUs) are corrected for Renilla signal and provided as percentages of wild-type eIF4A interaction with eIF4G(924–1444). These experiments were repeated three times in sextuplicate, and representative data are shown. The results are expressed as the mean ± standard deviation. (B) Immunoprecipitation of eIF4A/mutant-xpress with eIF4G(924–1444). (i) Two micrograms of HA antibody was added to 100 μg of RT101 cell lysates, which were then blotted with xpress antibody. (ii) Blots were stripped and probed with HA antibody to confirm equal G-Sepharose bead affinity for HA antibody. (iii) One-hundred microgram amounts of RT101 cell lysates were blotted with xpress antibody to show equal expression and loading of xpress-labeled protein. (iv) Densitometry readings were taken of each band in panel Bi, and the values were plotted against the RLU values in A.
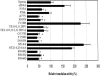
FIGURE 5.
The Q, I, Ia, GG, Ib, II, and VI conserved domains contribute to the translation-required regions of eIF4A. RT101 cells were transfected with 400 ng of wild-type (WT)/mutant eIF4A, 25 ng of stem–loop luciferase reporter, and 5 ng of thymidine kinase-Renilla luciferase gene. Values (relative light units) are corrected for Renilla signal and provided as percentages of wild-type eIF4A translational activity. These experiments were repeated three times in sextuplicate, and representative data are shown. The results are expressed as the mean ± standard deviation. (*) Significant difference compared with the control (xpress), as determined by the Student’s _t_-test (*,<0.0001).
FIGURE 6.
Pull-down of eIF4A/mutant-xpress with 7-methyl-GTP. (A) Thirty micrograms of cap 7-methyl-GTP Sepharose 4B were added to 100 μg of RT101 cell lysates transfected with eIF4A/mutant-xpress, spun down, and the SDS extract subjected to PAGE and immunoblotted with xpress antibody. Supernatant fractions, following spin-down of lysates with cap 7-methyl-GTP, were also blotted with xpress antibody. Prior to pull-down with 7-methyl-GTP, Sepharose 4B (50 μg) was added to 100 μg of RT101 cell lysates transfected with eIF4A/mutant-xpress, then blotted with xpress antibody. (B) Membranes were incubated with eIF4E antibody as a loading control for eIF4F complex.
FIGURE 7.
Wild-type but not eIF4A mutants inactivated for binding to Pdcd4 compete for interaction with eIF4GC. RT101 cells were transfected with 50 ng of pCMV-BD-eIF4G (924–1444), 400 ng of pCMV-AD-eIF4A or the mutants pCMV-AD-eIF4AF35A or pCMV-AD-eIF4AP56L, 25 ng of Gal4-luciferase reporter gene, and 5 ng of thymi-dine kinase-Renilla luciferase gene, with 0–200 ng of pCMV-Pdcd4. Values (relative light units) are corrected for Renilla signal and expressed as the mean ± standard deviation. Experiments were repeated three times in sextuplicate, and representative data are shown.
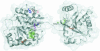
FIGURE 8.
Homology model of mouse eIF4A. A ribbon representation is superimposed on a transparent surface depiction of a homology model of mouse eIF4A. Specific amino acid residues are colored as follows: blue (F35, A77) = eIF4GM interaction; red (P56) = Pdcd4 interaction; purple (K83) = eIF4GM and Pdcd4 interaction; green (E112, L113, D183, R363) = eIF4GM and eIF4GC interaction; and orange (G137, T159, R360) = eIF4GM, eIF4GC, and Pdcd4 interaction. The figure was created using Pymol (
).
Similar articles
- Structure of the C-terminal MA-3 domain of the tumour suppressor protein Pdcd4 and characterization of its interaction with eIF4A.
Waters LC, Veverka V, Böhm M, Schmedt T, Choong PT, Muskett FW, Klempnauer KH, Carr MD. Waters LC, et al. Oncogene. 2007 Jul 26;26(34):4941-50. doi: 10.1038/sj.onc.1210305. Epub 2007 Feb 19. Oncogene. 2007. PMID: 17310995 - The transformation suppressor Pdcd4 is a novel eukaryotic translation initiation factor 4A binding protein that inhibits translation.
Yang HS, Jansen AP, Komar AA, Zheng X, Merrick WC, Costes S, Lockett SJ, Sonenberg N, Colburn NH. Yang HS, et al. Mol Cell Biol. 2003 Jan;23(1):26-37. doi: 10.1128/MCB.23.1.26-37.2003. Mol Cell Biol. 2003. PMID: 12482958 Free PMC article. - Leishmania infantum LeIF protein is an ATP-dependent RNA helicase and an eIF4A-like factor that inhibits translation in yeast.
Barhoumi M, Tanner NK, Banroques J, Linder P, Guizani I. Barhoumi M, et al. FEBS J. 2006 Nov;273(22):5086-100. doi: 10.1111/j.1742-4658.2006.05506.x. FEBS J. 2006. PMID: 17087726 - [Translational control by the poly(A) binding protein: a check for mRNA integrity].
Svitkin YV, Sonenberg N. Svitkin YV, et al. Mol Biol (Mosk). 2006 Jul-Aug;40(4):684-93. Mol Biol (Mosk). 2006. PMID: 16913227 Review. Russian. - The DEAD-box helicase eIF4A: paradigm or the odd one out?
Andreou AZ, Klostermeier D. Andreou AZ, et al. RNA Biol. 2013 Jan;10(1):19-32. doi: 10.4161/rna.21966. Epub 2012 Sep 20. RNA Biol. 2013. PMID: 22995829 Free PMC article. Review.
Cited by
- Pdcd4 repression of lysyl oxidase inhibits hypoxia-induced breast cancer cell invasion.
Santhanam AN, Baker AR, Hegamyer G, Kirschmann DA, Colburn NH. Santhanam AN, et al. Oncogene. 2010 Jul 8;29(27):3921-32. doi: 10.1038/onc.2010.158. Epub 2010 May 24. Oncogene. 2010. PMID: 20498644 Free PMC article. - Phylogenetic distribution and evolutionary history of bacterial DEAD-Box proteins.
López-Ramírez V, Alcaraz LD, Moreno-Hagelsieb G, Olmedo-Álvarez G. López-Ramírez V, et al. J Mol Evol. 2011 Apr;72(4):413-31. doi: 10.1007/s00239-011-9441-8. Epub 2011 Mar 25. J Mol Evol. 2011. PMID: 21437710 Free PMC article. - Programmed cell death 4 as an endogenous suppressor of BDNF translation is involved in stress-induced depression.
Li Y, Jia Y, Wang D, Zhuang X, Li Y, Guo C, Chu H, Zhu F, Wang J, Wang X, Wang Q, Zhao W, Shi Y, Chen W, Zhang L. Li Y, et al. Mol Psychiatry. 2021 Jun;26(6):2316-2333. doi: 10.1038/s41380-020-0692-x. Epub 2020 Mar 16. Mol Psychiatry. 2021. PMID: 32203159 Free PMC article. - The role of Pdcd4 in tumour suppression and protein translation.
Wang Q, Yang HS. Wang Q, et al. Biol Cell. 2018 May 28:10.1111/boc.201800014. doi: 10.1111/boc.201800014. Online ahead of print. Biol Cell. 2018. PMID: 29806708 Free PMC article. Review. - Tumorigenic activity and therapeutic inhibition of Rheb GTPase.
Mavrakis KJ, Zhu H, Silva RL, Mills JR, Teruya-Feldstein J, Lowe SW, Tam W, Pelletier J, Wendel HG. Mavrakis KJ, et al. Genes Dev. 2008 Aug 15;22(16):2178-88. doi: 10.1101/gad.1690808. Genes Dev. 2008. PMID: 18708578 Free PMC article.
References
- Afonja, O., Juste, D., Das, S., Matsuhashi, S., and Samuels, H.H. 2004. Induction of PDCD4 tumor suppressor gene expression by RAR agonists, antiestrogen and HER-2/neu antagonist in breast cancer cells. Evidence for a role in apoptosis. Oncogene 23: 8135–8145. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous