Novel avian influenza H7N3 strain outbreak, British Columbia - PubMed (original) (raw)
. 2004 Dec;10(12):2192-5.
doi: 10.3201/eid1012.040743.
Caroline R Astell, Malachi Griffith, Shaun M Coughlin, Michelle Moksa, Thomas Zeng, Duane E Smailus, Robert A Holt, Steven Jones, Marco A Marra, Martin Petric, Mel Krajden, David Lawrence, Annie Mak, Ron Chow, Danuta M Skowronski, S Aleina Tweed, SweeHan Goh, Robert C Brunham, John Robinson, Victoria Bowes, Ken Sojonky, Sean K Byrne, Yan Li, Darwyn Kobasa, Tim Booth, Mark Paetzel
Affiliations
- PMID: 15663859
- PMCID: PMC3323367
- DOI: 10.3201/eid1012.040743
Novel avian influenza H7N3 strain outbreak, British Columbia
Martin Hirst et al. Emerg Infect Dis. 2004 Dec.
Abstract
Genome sequences of chicken (low pathogenic avian influenza [LPAI] and highly pathogenic avian influenza [HPAI]) and human isolates from a 2004 outbreak of H7N3 avian influenza in Canada showed a novel insertion in the HA0 cleavage site of the human and HPAI isolate. This insertion likely occurred by recombination between the hemagglutination and matrix genes in the LPAI virus.
Figures
Figure 1
Alignment of the hemagglutinin cleavage region from four isolates of Fraser Valley H7N3 virus. A/Chicken/Canada/AVFV1/04 is designated AVFV1; A/Chicken/Canada/AVFV2/04 is designated AVFV2; A/Canada/444/04 (human) is Hu444, and A/Canada/504/04 (human) is Hu504). A 7–amino-acid (aa) insertion associated with the AVFV2 isolate and both human isolates is shown at aa 338.
Figure 2
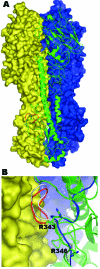
A homology model of the human A/Canada/504/04 (Hu504) hemagglutinin precursor (HA0) trimer based on the crystal structure of the human strain CV-1 HA0 (PDB: 1HA0) sequence identity 49.9%. A) Molecule A is shown as a green ribbon diagram; molecules B and C are shown in blue and yellow molecular surfaces, respectively. The 8–amino-acid (aa) sequence 335-342 (NPKQAYQK) is shown in red. B) A close up of this region located between molecules A (in green ribbon) and molecule C (in yellow surface). This 8-aa sequence forms a loop, which bumps into the adjoining molecule before energy minimization (gray). Shown in red is the loop after energy minimization, which results in the cleavage site's being pushed out slightly. Shown in blue is the corresponding region for the template structure (PDB code 1HA0). The side chains for arginine 343 and arginine 346 (–1 residue) are shown in stick form. (Since the preparation of this manuscript, the structure of an H7 HA protein has been reported [_12_]).
Figure A1
Complete nucleotide (A) and protein (B) alignments from the 5 isolates sequenced in this study. The alignments were generated by ClustalX (1.82) (
http://www.embl.de/\~chenna/clustal/darwin/
). [A/Canada/AVFV1/04 (environmental) is designated as chicken_A; A/Chicken/Canada/AVFV2/04 is designated as chicken_B; A/Canada/444/04 (human) is human_A and A/Canada/504/04 (human) is human_B. Full sequenced segments for chicken_D and chicken_E, not described in the text, are included for completeness.
Figure A2
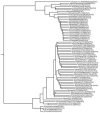
Phylogenetic tree of isolates described in the text with 65 full-length H7 HA gene segments present in GenBank. The tree was generated with nucleotide sequences by the neighbor-joining method in Bonsai (1.1.4.) (
http://calliope.gs.washington.edu/software/
). Corrected distance estimate is indicated by the scale bar. The H7N3 avian influenza isolates reported in this article are at the bottom of the tree.
Figure A3
A structure-based sequence alignment of the influenza A virus hemagglutinin precursor protein from human A/Canada/504/04 (Hu504) with that of the hemagglutinin precursor protein (HA0) from influenza A virus, human strain CV-1 whose structure was solved by Wiley and colleagues in 1998 (PDBcode 1HAO) (22). The alignment shows 49.9% identity in sequence. Insertions and deletions are highlighted in grey and nonconservative mutations are shown in yellow. Conserved cysteines are boxed. The glycosylated aparagines which are conserved are boxed with thin lines; the glycosylated asparagines that are not conserved are boxed with thick lines.
Similar articles
- Human illness and isolation of low-pathogenicity avian influenza virus of the H7N3 subtype in British Columbia, Canada.
Skowronski DM, Tweed SA, Petric M, Booth T, Li Y, Tam T. Skowronski DM, et al. J Infect Dis. 2006 Mar 15;193(6):899-900; author reply 900-1. doi: 10.1086/500219. J Infect Dis. 2006. PMID: 16479527 No abstract available. - Human illness from avian influenza H7N3, British Columbia.
Tweed SA, Skowronski DM, David ST, Larder A, Petric M, Lees W, Li Y, Katz J, Krajden M, Tellier R, Halpert C, Hirst M, Astell C, Lawrence D, Mak A. Tweed SA, et al. Emerg Infect Dis. 2004 Dec;10(12):2196-9. doi: 10.3201/eid1012.040961. Emerg Infect Dis. 2004. PMID: 15663860 Free PMC article. - Virus characterization, clinical presentation, and pathology associated with H7N3 avian influenza in British Columbia broiler breeder chickens in 2004.
Bowes VA, Ritchie SJ, Byrne S, Sojonky K, Bidulka JJ, Robinson JH. Bowes VA, et al. Avian Dis. 2004 Dec;48(4):928-34. doi: 10.1637/7218-060304R. Avian Dis. 2004. PMID: 15666877 - Avian influenza: the Canadian experience.
Pasick J, Berhane Y, Hooper-McGrevy K. Pasick J, et al. Rev Sci Tech. 2009 Apr;28(1):349-58. doi: 10.20506/rst.28.1.1875. Rev Sci Tech. 2009. PMID: 19618638 Review. - [Practical information on classical fowl plague (highly pathogenic avian influenza)].
Müller H. Müller H. Dtsch Tierarztl Wochenschr. 2003 Aug;110(8):330-2. Dtsch Tierarztl Wochenschr. 2003. PMID: 14535064 Review. German.
Cited by
- The first avian influenza A (H7N9) viral infection in humans in Zhejiang Province, China: a death report.
Chen E, Wang F, Lv H, Zhang Y, Ding H, Liu S, Cai J, Xie L, Xu X, Chai C, Mao H, Sun J, Lin J, Yu Z, Li L, Chen Z, Xia S. Chen E, et al. Front Med. 2013 Sep;7(3):333-44. doi: 10.1007/s11684-013-0275-1. Epub 2013 Jun 10. Front Med. 2013. PMID: 23757033 Free PMC article. - Genetic changes that accompanied shifts of low pathogenic avian influenza viruses toward higher pathogenicity in poultry.
Abdelwhab el-SM, Veits J, Mettenleiter TC. Abdelwhab el-SM, et al. Virulence. 2013 Aug 15;4(6):441-52. doi: 10.4161/viru.25710. Epub 2013 Jul 16. Virulence. 2013. PMID: 23863606 Free PMC article. Review. - Species-specific emergence of H7 highly pathogenic avian influenza virus is driven by intrahost selection differences between chickens and ducks.
de Bruin ACM, Spronken MI, Kok A, Rosu ME, de Meulder D, van Nieuwkoop S, Lexmond P, Funk M, Leijten LM, Bestebroer TM, Herfst S, van Riel D, Fouchier RAM, Richard M. de Bruin ACM, et al. PLoS Pathog. 2024 Feb 26;20(2):e1011942. doi: 10.1371/journal.ppat.1011942. eCollection 2024 Feb. PLoS Pathog. 2024. PMID: 38408092 Free PMC article. - Vaccines and antiviral drugs in pandemic preparedness.
Monto AS. Monto AS. Emerg Infect Dis. 2006 Jan;12(1):55-60. doi: 10.3201/eid1201.051068. Emerg Infect Dis. 2006. PMID: 16494718 Free PMC article. - Limited Antigenic Diversity in Contemporary H7 Avian-Origin Influenza A Viruses from North America.
Xu Y, Bailey E, Spackman E, Li T, Wang H, Long LP, Baroch JA, Cunningham FL, Lin X, Jarman RG, DeLiberto TJ, Wan XF. Xu Y, et al. Sci Rep. 2016 Feb 9;6:20688. doi: 10.1038/srep20688. Sci Rep. 2016. PMID: 26858078 Free PMC article.
References
- Perdue M, Crawford J, Garcia M, Latimer JE, Swayne D. Occurrence and possible mechanisms of cleavage site insertions in the avian influenza hemagglutinin gene. Swayne DE, Slemons RD, editors. Proceedings of the Fourth International Symposium on Avian Influenza. Kennett Square (PA): American Association of Avian Pathologists; 1998. p. 182–93.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical