Impaired intranuclear trafficking of Runx2 (AML3/CBFA1) transcription factors in breast cancer cells inhibits osteolysis in vivo - PubMed (original) (raw)
Impaired intranuclear trafficking of Runx2 (AML3/CBFA1) transcription factors in breast cancer cells inhibits osteolysis in vivo
Amjad Javed et al. Proc Natl Acad Sci U S A. 2005.
Abstract
Runx transcription factors comprise a family of proteins that are essential for organogenesis. A unique nuclear matrix-targeting signal in the C terminus directs these factors to their appropriate subnuclear domains. At these sites, they interact with coregulatory proteins and target genes. We have previously shown that aberrant expression of the Runx2 DNA binding domain in metastatic breast cancer cells can prevent production of osteolytic lesions in bone. Here, we show that proper Runx2 subnuclear targeting is required for osteolysis. We have identified point mutations of the Runx2 nuclear matrix-targeting signal sequence that impair its targeting to nuclear matrix sites. These mutations block the invasive and osteolytic properties of MDA-MB-231 breast cancer cells in vivo. Cell lines expressing this Runx2 mutant protein inhibit the osteogenic properties of bone marrow stromal cells in coculture assays. The mutant breast cancer cells also exhibit reduced invasiveness in vitro and do not express genes involved in invasion and angiogenesis (VEGF and MMP13). Our findings suggest that fidelity of Runx2 intranuclear organization is necessary for expression of target genes that mediate the osteolytic activity of metastatic breast cancer cells.
Figures
Fig. 1.
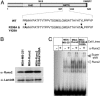
WT and R398A and Y428A mutant Runx2 proteins are expressed in MDA-MB-231 stable cells and exhibit similar DNA binding activity. (A) Schematic representation of the mouse Runx2 protein with key domains highlighted (RHD, DNA binding runt homology domain; NLS, nuclear localization signal; NMTS; and VWRPY, a penta-peptide motif conserved among Runx factors). At the bottom, the 38 amino-acids (397–434) that constitute the NMTS are shown. Arginine and tyrosine at positions 398 and 428, respectively, were mutated to alanine. (B) MDA-MB-231 breast cancer cells were stably transfected with either WT or R398A and Y428A mutant Runx2 expression plasmids. Cell lysates from parental and stable lines were resolved in 10% SDS/PAGE, and Western blots were probed with mouse Runx2 mAb (44). Lamin B antigen is shown as a loading control. (C) Electrophoretic mobility shift analysis of Runx2 DNA binding activity from breast cancer lines. Nuclear extracts were isolated from the indicated cell lines as described (18). Runx consensus oligonucleotide used as probe was mixed with 10 μg of nuclear extract in the presence (+) or absence (-) of Runx2 Ab. Arrowhead indicates the Runx2–DNA complex.
Fig. 2.
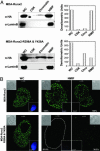
Paired mutation of R398A and Y428A in Runx2 protein results in loss of nuclear matrix association in breast cancer cells. (A) Biochemical fractionation of breast cancer lines expressing WT or targeting-deficient Runx2 mutant protein. Harvested cell pellets were subjected to high-salt and detergent extraction to isolate CSK, chromatin, and NMIF fractions, as described in Materials and Methods. Equal fraction volumes based on cell equivalence were resolved in 10% SDS/PAGE, and membranes were probed with mouse HA mAb to detect Runx2 protein (Left). Integrity of subcellular fractions is confirmed by Lamin B, a marker for nuclear matrix. Right shows graphic representations of densitometric quantitation of Western blotting signals. (B) In situ immunofluorescence microscopy of Runx2 proteins in breast cancer stable lines. Cells were cultured on gelatin-coated cover slips and processed as WC or extracted to reveal the NMIF. Two representative cells are shown for WT (Upper) and R398A and Y428A Runx2 mutant (Lower). Cells from four independent NMIF preparations were counted, and percentages of HA-stained cells are shown. Insets show differential interference contrast (DIC) or DAPI stained images of the cell. Removal of chromatin in the NMIF preparations is reflected by the absence of DAPI staining. Magnifications are ×100.
Fig. 3.
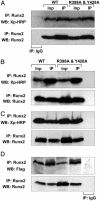
Coregulatory protein interactions across the targeting-deficient Runx2 mutant protein are preserved. HeLa cells that do not express endogenous Runx proteins were transiently cotransfected with 5 μg of expression plasmid for WT or mutant Runx2 and coregulatory proteins: Groucho/TLE, a C terminus interacting protein (A); Cbfβ, an N terminus interacting protein (B); and two proteins that interact at the NMTS domain, Homeodomain Msx2 (C) and BMP2 mediator Smad1 (D). Cells were collected 24 h later, and immunoprecipitations were performed with Runx2 Ab. Immunocomplexes were resolved in 10–12% SDS/PAGE, and Western blots were probed as indicated. Inp, 10% of input control.
Fig. 4.
Inhibition of bone marrow cell osteogenic differentiation by breast cancer cells is linked to subnuclear targeting of Runx2. Breast cancer cells (5,000) expressing WT or mutant Runx2 were added to 5-day-old murine BMSC cultures. Adherent cells were cultured for an additional 16 days in osteogenic medium (α-MEM supplemented with 10% FBS/10-8 M dexamethasone/50 μg/ml ascorbic acid/8 mM β-glycerol phosphate). (A) Plates were then stained by Von Kossa to reveal mineralized nodules. Three representative wells for each group are shown. (B) RPA of murine osteoblast marker gene expression in cocultures of mouse BMSC and human breast cancer cells. Total RNA was isolated from 20-day coculture cells and probed for osteoblastic markers as described in Materials and Methods. L, Undigested probe ladder; 1, BMSC alone; 2, BMSC cocultured with MDA-MB-231 cells; 3, BMSC cocultured with MDA cells stably expressing mutant Runx2. Key markers are osteopontin (OPN), bone sialoprotein (BSP), collagen type I (COL I), osteocalcin (OC), and ribosomal protein gene (L32) as an internal control.
Fig. 5.
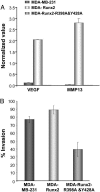
Compromised Runx2 subnuclear targeting modifies phenotypic properties of the MDA breast cancer cells. RNA was isolated from MDA stable lines and osteogenic gene profiling assessed as described in Materials and Methods. (A) Validation of modified genes by quantitative RT-PCR. Changes in expression of two selected Runx2 responsive genes (VEGF and MMP13) were confirmed by real-time PCR. Values were normalized to GAPDH, which was equally expressed in the three cell lines. Significant induction of both genes was observed only in breast cancer stable lines expressing WT Runx2. (B) Runx2 regulates invasive potential of breast cancer cells. Cells from parental MDA-MB-231 or WT and mutant Runx2 stable breast cancer lines were plated in chambers as described. The Matrigel layer was then removed and the number of migrated cells counted. Data are plotted as percentage of cells that migrated through Matrigel in comparison with membrane control alone.
Fig. 6.
Subnuclear trafficking of Runx2 is associated with in vivo formation of osteolytic lesions by breast cancer cells. (A) Control (no cell), MDA-MB-231 parental cell, or targeting-deficient mutant Runx2 (R398A & Y428A) expressing stable cells were injected into the intramedullary region of the tibia of 4- to 6-week-old SCID mice. Formation of osteolytic lesions in bones was assessed by radiography (see text). (B) RPA of genes reflecting bone resorbing activity. Total RNA from mouse BMSC and human breast cancer cell cocultures was isolated and probed for mouse osteoclast-related markers, as described in Materials and Methods. Markers are as follows: inducible nitrous oxide (iNOS), TNF-α κβ ligand (RANKL), IL-6, tartrate-resistant acid phosphatase (TRAP) and the internal control ribosomal protein gene (L32). Increased expression of WT Runx2 does not significantly modify gene expression in the parental MDA-MB-231 cells.
Similar articles
- The Runx2 osteogenic transcription factor regulates matrix metalloproteinase 9 in bone metastatic cancer cells and controls cell invasion.
Pratap J, Javed A, Languino LR, van Wijnen AJ, Stein JL, Stein GS, Lian JB. Pratap J, et al. Mol Cell Biol. 2005 Oct;25(19):8581-91. doi: 10.1128/MCB.25.19.8581-8591.2005. Mol Cell Biol. 2005. PMID: 16166639 Free PMC article. - Smad3 interacts with JunB and Cbfa1/Runx2 for transforming growth factor-beta1-stimulated collagenase-3 expression in human breast cancer cells.
Selvamurugan N, Kwok S, Partridge NC. Selvamurugan N, et al. J Biol Chem. 2004 Jun 25;279(26):27764-73. doi: 10.1074/jbc.M312870200. Epub 2004 Apr 14. J Biol Chem. 2004. PMID: 15084595 - Runx2 induces bone osteolysis by transcriptional suppression of TSSC1.
Wang DC, Wang HF, Yuan ZN. Wang DC, et al. Biochem Biophys Res Commun. 2013 Sep 6;438(4):635-9. doi: 10.1016/j.bbrc.2013.07.131. Epub 2013 Aug 8. Biochem Biophys Res Commun. 2013. PMID: 23933319 - Temporal and spatial parameters of skeletal gene expression: targeting RUNX factors and their coregulatory proteins to subnuclear domains.
Stein GS, Lian JB, Stein JL, van Wijnen AJ, Choi JY, Pratap J, Zaidi SK. Stein GS, et al. Connect Tissue Res. 2003;44 Suppl 1:149-53. Connect Tissue Res. 2003. PMID: 12952189 Review. - Regulatory controls for osteoblast growth and differentiation: role of Runx/Cbfa/AML factors.
Lian JB, Javed A, Zaidi SK, Lengner C, Montecino M, van Wijnen AJ, Stein JL, Stein GS. Lian JB, et al. Crit Rev Eukaryot Gene Expr. 2004;14(1-2):1-41. Crit Rev Eukaryot Gene Expr. 2004. PMID: 15104525 Review.
Cited by
- YTHDF2-mediated FGF14-AS2 decay promotes osteolytic metastasis of breast cancer by enhancing RUNX2 mRNA translation.
Zhang M, Wang J, Jin Y, Zheng Q, Xing M, Tang Y, Ma Y, Li L, Yao B, Wu H, Ma C. Zhang M, et al. Br J Cancer. 2022 Dec;127(12):2141-2153. doi: 10.1038/s41416-022-02006-y. Epub 2022 Oct 10. Br J Cancer. 2022. PMID: 36216883 Free PMC article. - Tumor Suppressor WWOX inhibits osteosarcoma metastasis by modulating RUNX2 function.
Del Mare S, Aqeilan RI. Del Mare S, et al. Sci Rep. 2015 Aug 10;5:12959. doi: 10.1038/srep12959. Sci Rep. 2015. PMID: 26256646 Free PMC article. - Definitive hematopoiesis requires Runx1 C-terminal-mediated subnuclear targeting and transactivation.
Dowdy CR, Xie R, Frederick D, Hussain S, Zaidi SK, Vradii D, Javed A, Li X, Jones SN, Lian JB, van Wijnen AJ, Stein JL, Stein GS. Dowdy CR, et al. Hum Mol Genet. 2010 Mar 15;19(6):1048-57. doi: 10.1093/hmg/ddp568. Epub 2009 Dec 24. Hum Mol Genet. 2010. PMID: 20035012 Free PMC article. - Runx2 transcriptional activation of Indian Hedgehog and a downstream bone metastatic pathway in breast cancer cells.
Pratap J, Wixted JJ, Gaur T, Zaidi SK, Dobson J, Gokul KD, Hussain S, van Wijnen AJ, Stein JL, Stein GS, Lian JB. Pratap J, et al. Cancer Res. 2008 Oct 1;68(19):7795-802. doi: 10.1158/0008-5472.CAN-08-1078. Cancer Res. 2008. PMID: 18829534 Free PMC article. - RUNX3 expression is lost in glioma and its restoration causes drastic suppression of tumor invasion and migration.
Mei PJ, Bai J, Liu H, Li C, Wu YP, Yu ZQ, Zheng JN. Mei PJ, et al. J Cancer Res Clin Oncol. 2011 Dec;137(12):1823-30. doi: 10.1007/s00432-011-1063-4. Epub 2011 Sep 16. J Cancer Res Clin Oncol. 2011. PMID: 21922326
References
- Okuda, T., van Deursen, J., Hiebert, S. W., Grosveld, G. & Downing, J. R. (1996) Cell 84, 321-330. - PubMed
- Komori, T., Yagi, H., Nomura, S., Yamaguchi, A., Sasaki, K., Deguchi, K., Shimizu, Y., Bronson, R. T., Gao, Y.-H., Inada, M., et al. (1997) Cell 89, 755-764. - PubMed
- Li, Q. L., Ito, K., Sakakura, C., Fukamachi, H., Inoue, K., Chi, X. Z., Lee, K. Y., Nomura, S., Lee, C. W., Han, S. B., et al. (2002) Cell 109, 113-124. - PubMed
- Nucifora, G. & Rowley, J. D. (1995) Blood 86, 1-14. - PubMed
- Mundlos, S., Otto, F., Mundlos, C., Mulliken, J. B., Aylsworth, A. S., Albright, S., Lindhout, D., Cole, W. G., Henn, W., Knoll, J. H. M., et al. (1997) Cell 89, 773-779. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P01 AR048818/AR/NIAMS NIH HHS/United States
- R01AR47045/AR/NIAMS NIH HHS/United States
- R01 AR047045/AR/NIAMS NIH HHS/United States
- P30 DK032520/DK/NIDDK NIH HHS/United States
- DK32520/DK/NIDDK NIH HHS/United States
- P01CA82834/CA/NCI NIH HHS/United States
- R01AR047045/AR/NIAMS NIH HHS/United States
- P01 CA082834/CA/NCI NIH HHS/United States
- P01AR0409920/AR/NIAMS NIH HHS/United States
- P01AR48818/AR/NIAMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous