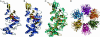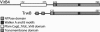Predicted hexameric structure of the Agrobacterium VirB4 C terminus suggests VirB4 acts as a docking site during type IV secretion - PubMed (original) (raw)
Comparative Study
. 2005 Feb 1;102(5):1685-90.
doi: 10.1073/pnas.0409399102. Epub 2005 Jan 24.
Affiliations
- PMID: 15668378
- PMCID: PMC547840
- DOI: 10.1073/pnas.0409399102
Comparative Study
Predicted hexameric structure of the Agrobacterium VirB4 C terminus suggests VirB4 acts as a docking site during type IV secretion
Rebecca Middleton et al. Proc Natl Acad Sci U S A. 2005.
Abstract
The Agrobacterium T-DNA transporter belongs to a growing class of evolutionarily conserved transporters, called type IV secretion systems (T4SSs). VirB4, 789 aa, is the largest T4SS component, providing a rich source of possible structural domains. Here, we use a variety of bioinformatics methods to predict that the C-terminal domain of VirB4 (including the Walker A and B nucleotide-binding motifs) is related by divergent evolution to the cytoplasmic domain of TrwB, the coupling protein required for conjugative transfer of plasmid R388 from Escherichia coli. This prediction is supported by detailed sequence and structure analyses showing conservation of functionally and structurally important residues between VirB4 and TrwB. The availability of a solved crystal structure for TrwB enables the construction of a comparative model for VirB4 and the prediction that, like TrwB, VirB4 forms a hexamer. These results lead to a model in which VirB4 acts as a docking site at the entrance of the T4SS channel and acts in concert with VirD4 and VirB11 to transport substrates (T-strand linked to VirD2 or proteins such as VirE2, VirE3, or VirF) through the T4SS.
Figures
Fig. 4.
MSA of VirB4 and TrwB and selected homologs along their NTP-binding domains. Sequence homologs to VirB4 are shown directly below VirB4, followed by TrwB and homologs. Walker A residues are boxed in red, Walker B residues are boxed in magenta, and the conserved glutamine is highlighted in green.
Fig. 2.
VirB4-TrwB pairwise alignment. The cytoplasmic domain of TrwB (residues 122–507) and the C terminus of VirB4 (residues 425–789) are aligned, with identical and similar residues shown in black and gray, respectively. Boxed areas highlight the Walker A and B motifs. The Walker A motif is conserved across VirB4 and TrwB (VirB4, G433-T440; TrwB, G130-S137). The Walker B motif is found in its entirety in VirB4 (R619-E635), whereas a truncated version is evident in TrwB (R349-E357).
Fig. 3.
Homology model for the C terminus of VirB4. (A) VirB4 monomer model. α-helices are blue and β-sheets are gold. The Walker A motif (G433-T440) is red, the Walker B motif (R619-E635) is magenta, and the conserved Q668 is green. (B) Monomer of TrwB, using the same coloring as for VirB4. (C) Superposition of VIRB4 (turquoise) and TrwB (gold). (D) Predicted hexameric structure of VirB4.
Fig. 1.
Domain organization of VirB4 and TrwB. VirB4 and TrwB share a C-terminal NTP-binding domain but have different N termini. The N terminus of VirB4 matches the
pfam
domain CagE_TrbE_VirB, but has no known function or structure, whereas the TrwB N terminus contains two predicted TM helices.
Fig. 5.
Surface image of VirB4 monomer, highlighting the NTP-binding cleft. To better display the NTP-binding cleft, we show the structure from a different perspective, corresponding to the back side of Fig. 3_A_. The conserved lysine (K) of the Walker A motif is red, the conserved DE of the Walker B motif is purple, and the conserved glutamine (Q) is highlighted in yellow.
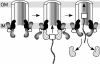
Fig. 6.
Model for the function of VirB4 during the type IV secretion process. VirB4 is shaded in gray as two domains linked by a hinge (solid line). VirB11 and VirD4 are black and stippled, respectively. For simplicity, only two subunits of VirB4, VirD4, and VirB11 are shown, and the rest of the T4SS is drawn as a half cylinder. VirD2 is shown as a white square attached to the T-strand (wave ladder-like line). Arrows indicate subunit exchange between VirB4 and VirD4. See text for details. OM, outer membrane; IM, inner membrane.
Similar articles
- The All-Alpha Domains of Coupling Proteins from the Agrobacterium tumefaciens VirB/VirD4 and Enterococcus faecalis pCF10-Encoded Type IV Secretion Systems Confer Specificity to Binding of Cognate DNA Substrates.
Whitaker N, Chen Y, Jakubowski SJ, Sarkar MK, Li F, Christie PJ. Whitaker N, et al. J Bacteriol. 2015 Jul;197(14):2335-49. doi: 10.1128/JB.00189-15. Epub 2015 May 4. J Bacteriol. 2015. PMID: 25939830 Free PMC article. - In Situ Visualization of the pKM101-Encoded Type IV Secretion System Reveals a Highly Symmetric ATPase Energy Center.
Khara P, Song L, Christie PJ, Hu B. Khara P, et al. mBio. 2021 Oct 26;12(5):e0246521. doi: 10.1128/mBio.02465-21. Epub 2021 Oct 12. mBio. 2021. PMID: 34634937 Free PMC article. - Agrobacterium-mediated T-DNA transfer and integration by minimal VirD2 consisting of the relaxase domain and a type IV secretion system translocation signal.
van Kregten M, Lindhout BI, Hooykaas PJ, van der Zaal BJ. van Kregten M, et al. Mol Plant Microbe Interact. 2009 Nov;22(11):1356-65. doi: 10.1094/MPMI-22-11-1356. Mol Plant Microbe Interact. 2009. PMID: 19810805 - A molecular understanding of the catalytic cycle of the nucleotide-binding domain of the ABC transporter HlyB.
Zaitseva J, Jenewein S, Oswald C, Jumpertz T, Holland IB, Schmitt L. Zaitseva J, et al. Biochem Soc Trans. 2005 Nov;33(Pt 5):990-5. doi: 10.1042/BST20050990. Biochem Soc Trans. 2005. PMID: 16246029 Review. - Exploring cargo transport mechanics in the type IV secretion systems.
Li J, Wolf SG, Elbaum M, Tzfira T. Li J, et al. Trends Microbiol. 2005 Jul;13(7):295-8. doi: 10.1016/j.tim.2005.05.002. Trends Microbiol. 2005. PMID: 15923116 Review.
Cited by
- Structural insights into the membrane-extracted dimeric form of the ATPase TraB from the Escherichia coli pKM101 conjugation system.
Durand E, Waksman G, Receveur-Brechot V. Durand E, et al. BMC Struct Biol. 2011 Jan 25;11:4. doi: 10.1186/1472-6807-11-4. BMC Struct Biol. 2011. PMID: 21266026 Free PMC article. - Agrobacterium tumefaciens VirB8 structure reveals potential protein-protein interaction sites.
Bailey S, Ward D, Middleton R, Grossmann JG, Zambryski PC. Bailey S, et al. Proc Natl Acad Sci U S A. 2006 Feb 21;103(8):2582-7. doi: 10.1073/pnas.0511216103. Epub 2006 Feb 15. Proc Natl Acad Sci U S A. 2006. PMID: 16481621 Free PMC article. - ATPase activity and oligomeric state of TrwK, the VirB4 homologue of the plasmid R388 type IV secretion system.
Arechaga I, Peña A, Zunzunegui S, del Carmen Fernández-Alonso M, Rivas G, de la Cruz F. Arechaga I, et al. J Bacteriol. 2008 Aug;190(15):5472-9. doi: 10.1128/JB.00321-08. Epub 2008 Jun 6. J Bacteriol. 2008. PMID: 18539740 Free PMC article. - Biological diversity of prokaryotic type IV secretion systems.
Alvarez-Martinez CE, Christie PJ. Alvarez-Martinez CE, et al. Microbiol Mol Biol Rev. 2009 Dec;73(4):775-808. doi: 10.1128/MMBR.00023-09. Microbiol Mol Biol Rev. 2009. PMID: 19946141 Free PMC article. Review. - GI-type T4SS-mediated horizontal transfer of the 89K pathogenicity island in epidemic Streptococcus suis serotype 2.
Li M, Shen X, Yan J, Han H, Zheng B, Liu D, Cheng H, Zhao Y, Rao X, Wang C, Tang J, Hu F, Gao GF. Li M, et al. Mol Microbiol. 2011 Mar;79(6):1670-83. doi: 10.1111/j.1365-2958.2011.07553.x. Epub 2011 Feb 10. Mol Microbiol. 2011. PMID: 21244532 Free PMC article.
References
- Baron, C., O'Callaghan, D. & Lanka, E. (2002) Mol. Microbiol. 43, 1359–1365. - PubMed
- Cao, T. B. & Saier, M. H., Jr. (2001) Microbiology 147, 3201–3214. - PubMed
- Sexton, J. A. & Vogel, J. P. (2002) Traffic 3, 178–185. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources