Use of pyrosequencing of 16S rRNA fragments to differentiate between bacteria responsible for neonatal sepsis - PubMed (original) (raw)
Use of pyrosequencing of 16S rRNA fragments to differentiate between bacteria responsible for neonatal sepsis
Jeanne A Jordan et al. J Mol Diagn. 2005 Feb.
Abstract
Infants admitted to neonatal intensive care units for suspicion of bacterial sepsis receive at least two broad-spectrum antibiotics for a minimum of 48 to 72 hours to cover both gram-positive and gram-negative organisms while awaiting blood culture results. On average, bacterial growth becomes detectable within 12 to 24 hours, with an additional 24 to 48 hours required for identification. We have previously described using a 16S rRNA PCR assay for screening neonatal blood for bacterial DNA. Combining PCR with DNA sequencing could prove a faster means of detecting bacteria than culture-based identification. If successful, antibiotic therapy could be appropriately tailored sooner, thus sparing infants the administration of unnecessary antibiotics. Our goal was to assess the potential of pyrosequencing to differentiate between bacteria commonly associated with neonatal sepsis. To begin, full-length sequencing of the 380-bp 16S rRNA amplicons from representative bacteria was conducted (ABI 3100) and several databases queried. These included Staphylococcus sp., Streptococcus sp., Listeria sp., and numerous gram-negative rods. The sequences from clinical isolates were identical to those present in the published databases for the same bacteria. As a result, an informative 15 bases within the 380-bp amplicon was targeted for pyrosequencing following enrichment culture and PCR amplification. A total of 643 bacterial isolates commonly associated with neonatal sepsis, and 15 PCR-positive, culture-positive neonatal whole blood samples were analyzed by pyrosequencing. Results of DNA sequencing and culture identification were compared. In summary, we were successful at using PCR and pyrosequencing together to accurately differentiate between highly diverse bacterial groups.
Figures
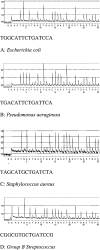
Figure 1
A–D: Representative pyrograms and the informative 15 base molecular fingerprint generated from representative bacteria associated with neonatal sepsis.
Similar articles
- Direct Screening of Blood by PCR and Pyrosequencing for a 16S rRNA Gene Target from Emergency Department and Intensive Care Unit Patients Being Evaluated for Bloodstream Infection.
Moore MS, McCarroll MG, McCann CD, May L, Younes N, Jordan JA. Moore MS, et al. J Clin Microbiol. 2016 Jan;54(1):99-105. doi: 10.1128/JCM.02394-15. Epub 2015 Oct 28. J Clin Microbiol. 2016. PMID: 26511737 Free PMC article. - Detection of bacterial DNA by PCR and reverse hybridization in the 16S rRNA gene with particular reference to neonatal septicemia.
Shang S, Chen Z, Yu X. Shang S, et al. Acta Paediatr. 2001 Feb;90(2):179-83. doi: 10.1080/080352501300049389. Acta Paediatr. 2001. PMID: 11236048 - A molecular gram stain using broad range PCR and pyrosequencing technology: a potentially useful tool for diagnosing orthopaedic infections.
Kobayashi N, Bauer TW, Togawa D, Lieberman IH, Sakai H, Fujishiro T, Tuohy MJ, Procop GW. Kobayashi N, et al. Diagn Mol Pathol. 2005 Jun;14(2):83-9. doi: 10.1097/01.pas.0000162753.38284.1a. Diagn Mol Pathol. 2005. PMID: 15905691 - Utility of pyrosequencing in identifying bacteria directly from positive blood culture bottles.
Jordan JA, Jones-Laughner J, Durso MB. Jordan JA, et al. J Clin Microbiol. 2009 Feb;47(2):368-72. doi: 10.1128/JCM.01991-08. Epub 2008 Dec 17. J Clin Microbiol. 2009. PMID: 19091813 Free PMC article. - Literature review on the distribution characteristics and antimicrobial resistance of bacterial pathogens in neonatal sepsis.
Wang J, Zhang H, Yan J, Zhang T. Wang J, et al. J Matern Fetal Neonatal Med. 2022 Mar;35(5):861-870. doi: 10.1080/14767058.2020.1732342. Epub 2020 Feb 26. J Matern Fetal Neonatal Med. 2022. PMID: 32102584 Review.
Cited by
- DNAemia detection by multiplex PCR and biomarkers for infection in systemic inflammatory response syndrome patients.
Fitting C, Parlato M, Adib-Conquy M, Memain N, Philippart F, Misset B, Monchi M, Cavaillon JM, Adrie C. Fitting C, et al. PLoS One. 2012;7(6):e38916. doi: 10.1371/journal.pone.0038916. Epub 2012 Jun 15. PLoS One. 2012. PMID: 22719987 Free PMC article. - Call for a quality standard for sequence-based assays in clinical microbiology: necessity for quality assessment of sequences used in microbial identification and typing.
Underwood A, Green J. Underwood A, et al. J Clin Microbiol. 2011 Jan;49(1):23-6. doi: 10.1128/JCM.01918-10. Epub 2010 Nov 10. J Clin Microbiol. 2011. PMID: 21068275 Free PMC article. Review. No abstract available. - Detection of EGFR and KRAS Mutation by Pyrosequencing Analysis in Cytologic Samples of Non-Small Cell Lung Cancer.
Lee SE, Lee SY, Park HK, Oh SY, Kim HJ, Lee KY, Kim WS. Lee SE, et al. J Korean Med Sci. 2016 Aug;31(8):1224-30. doi: 10.3346/jkms.2016.31.8.1224. Epub 2016 May 12. J Korean Med Sci. 2016. PMID: 27478332 Free PMC article. - Identification of human pathogens isolated from blood using microarray hybridisation and signal pattern recognition.
Wiesinger-Mayr H, Vierlinger K, Pichler R, Kriegner A, Hirschl AM, Presterl E, Bodrossy L, Noehammer C. Wiesinger-Mayr H, et al. BMC Microbiol. 2007 Aug 14;7:78. doi: 10.1186/1471-2180-7-78. BMC Microbiol. 2007. PMID: 17697354 Free PMC article. - Pyrosequencing for mini-barcoding of fresh and old museum specimens.
Shokralla S, Zhou X, Janzen DH, Hallwachs W, Landry JF, Jacobus LM, Hajibabaei M. Shokralla S, et al. PLoS One. 2011;6(7):e21252. doi: 10.1371/journal.pone.0021252. Epub 2011 Jul 27. PLoS One. 2011. PMID: 21818256 Free PMC article.
References
- Cerase PA. Neonatal sepsis. J Perinatal Neonatal Nursing. 1989;3:48–57. - PubMed
- Gerdes JS. Clinicopathologic approach to the diagnosis of neonatal sepsis. Clin Perinatol. 1991;18:361–381. - PubMed
- Klein JO, Marcy SM. Bacterial sepsis and meningitis. Infectious Diseases of the Fetus and Newborn. Remington J, Klein J, editors. Philadelphia: W.B. Saunders; 1990:p 601.
- Witek-Janusek L, Cusack C. Neonatal sepsis: confronting the challenge. Crit Care Nurs Clin North Am. 1994;6:405–419. - PubMed
- Freedman RM, Ingram DL, Gross I, Ehrenkranz RA, Warshaw JB, Baltimore RS. A half century of neonatal sepsis at Yale: 1928 to 1978. Am J Dis Child. 1981;135:140–144. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials