PR-Set7-dependent methylation of histone H4 Lys 20 functions in repression of gene expression and is essential for mitosis - PubMed (original) (raw)
. 2005 Feb 15;19(4):431-5.
doi: 10.1101/gad.1263005. Epub 2005 Jan 28.
Affiliations
- PMID: 15681608
- PMCID: PMC548943
- DOI: 10.1101/gad.1263005
PR-Set7-dependent methylation of histone H4 Lys 20 functions in repression of gene expression and is essential for mitosis
Dmitry Karachentsev et al. Genes Dev. 2005.
Abstract
The histone methyl transferase PR-Set7 mediates histone H4 Lys 20 methylation, a mark of constitutive and facultative heterochromatin. We isolated a null mutation in Drosophila PR-Set7 that suppresses position effect variegation, indicating that PR-Set7 indeed functions in silencing general gene expression. In PR-Set7 larval leg and eye discs, the number of cells is lower than normal, and the DNA content in these cells is significantly increased. These data show that PR-Set7-dependent methylation is essential for the process of mitosis. The methylation mark is highly stable and is maintained even in the absence of PR-Set7 protein.
Figures
Figure 1.
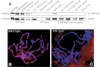
PR-Set720 mutants lack the PR-Set7 methylase. (A) Western blot of protein extracts derived from wild-type and homozygous PR-Set720 tissue, probed with anti-Drosophila PR-Set7 antibodies. (B,C) Immunofluorescent staining of wild-type (B) and PR-Set720 (C) salivary gland chromosomes with anti-Drosophila PR-Set7 antibodies (red) and Hoechst DNA dye (blue).
Figure 2.
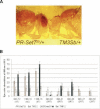
Reduction of PR-Set7 suppresses position effect variegation (PEV). (A, right) TM3Sb/+ fly showing variegated expression of a w+ transgene. (Left) PR-Set720/+ fly has red eyes because the variegated expression of the w+ transgene has been suppressed. (B) Measurement of eye-color (OD 480 nm) of extracts from different transgenic w+ lines expression, in heterozygous PR-Set7 flies (PR-Set720/+, or Df(3R)red31/+), or wild-type flies (Bal/+). Deletion of one copy of the PR-Set7 gene increases the eye color significantly, confirming the suppression of variegation. The balancers used were TM3 and TM6. The transgenic lines used were 36C-72 and 118E-15 containing w+ in the telomeric heterochromatin of the fourth chromosome. 118E-10 contains a w+ transgene in the centromeric heterochromatin. All second and third w+ transgenic lines contain the transgene in the telomeric heterochromatin.
Figure 3.
The mono-, di-, and trimethylation mark of H4-K20 is reduced in PR-Set7 mutants. Immunofluorescent staining of wild-type (A,C,E) and PR-Set720 (B,D,F) salivary gland chromosomes with anti-monomethylated (A,B), anti-dimethylated (C,D), and anti-trimethylated (E,F) H4-K20 antibodies (red) and antibody recognizing the catalytic subunit of RNA polymerase II (green). DNA dye Hoechst (blue).
Figure 4.
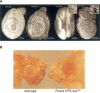
In PR-Set720 animals, the mitotic process is interrupted. (A) Leg imaginal discs (left) and eye imaginal discs (right) of wild-type and PR-Set7 larvae stained with the DNA dye Hoechst (white). (B) Eyes of wild-type and PR-Set7 partial loss-of-function flies. Note the smaller eye size in irregular organization of the mutant eye.
Similar articles
- Mitotic-specific methylation of histone H4 Lys 20 follows increased PR-Set7 expression and its localization to mitotic chromosomes.
Rice JC, Nishioka K, Sarma K, Steward R, Reinberg D, Allis CD. Rice JC, et al. Genes Dev. 2002 Sep 1;16(17):2225-30. doi: 10.1101/gad.1014902. Genes Dev. 2002. PMID: 12208845 Free PMC article. - Methylation of histone H4 lysine 20 by PR-Set7 ensures the integrity of late replicating sequence domains in Drosophila.
Li Y, Armstrong RL, Duronio RJ, MacAlpine DM. Li Y, et al. Nucleic Acids Res. 2016 Sep 6;44(15):7204-18. doi: 10.1093/nar/gkw333. Epub 2016 Apr 29. Nucleic Acids Res. 2016. PMID: 27131378 Free PMC article. - PR-Set7 is a nucleosome-specific methyltransferase that modifies lysine 20 of histone H4 and is associated with silent chromatin.
Nishioka K, Rice JC, Sarma K, Erdjument-Bromage H, Werner J, Wang Y, Chuikov S, Valenzuela P, Tempst P, Steward R, Lis JT, Allis CD, Reinberg D. Nishioka K, et al. Mol Cell. 2002 Jun;9(6):1201-13. doi: 10.1016/s1097-2765(02)00548-8. Mol Cell. 2002. PMID: 12086618 - The diverse functions of histone lysine methylation.
Martin C, Zhang Y. Martin C, et al. Nat Rev Mol Cell Biol. 2005 Nov;6(11):838-49. doi: 10.1038/nrm1761. Nat Rev Mol Cell Biol. 2005. PMID: 16261189 Review. - The multiple facets of histone H4-lysine 20 methylation.
Yang H, Mizzen CA. Yang H, et al. Biochem Cell Biol. 2009 Feb;87(1):151-61. doi: 10.1139/O08-131. Biochem Cell Biol. 2009. PMID: 19234531 Review.
Cited by
- Histone H4 acetylation differentially modulates proliferation in adult oligodendrocyte progenitors.
Dansu DK, Selcen I, Sauma S, Prentice E, Huang D, Li M, Moyon S, Casaccia P. Dansu DK, et al. J Cell Biol. 2024 Nov 4;223(11):e202308064. doi: 10.1083/jcb.202308064. Epub 2024 Aug 12. J Cell Biol. 2024. PMID: 39133301 - Drosophila melanogaster Set8 and L(3)mbt function in gene expression independently of histone H4 lysine 20 methylation.
Crain AT, Butler MB, Hill CA, Huynh M, McGinty RK, Duronio RJ. Crain AT, et al. Genes Dev. 2024 Jun 25;38(9-10):455-472. doi: 10.1101/gad.351698.124. Genes Dev. 2024. PMID: 38866557 Free PMC article. - Mono-methylated histones control PARP-1 in chromatin and transcription.
Bamgbose G, Bordet G, Lodhi N, Tulin A. Bamgbose G, et al. Elife. 2024 May 1;13:RP91482. doi: 10.7554/eLife.91482. Elife. 2024. PMID: 38690995 Free PMC article. - MoCoLo: a testing framework for motif co-localization.
Xu Q, Del Mundo IMA, Zewail-Foote M, Luke BT, Vasquez KM, Kowalski J. Xu Q, et al. Brief Bioinform. 2024 Jan 22;25(2):bbae019. doi: 10.1093/bib/bbae019. Brief Bioinform. 2024. PMID: 38521050 Free PMC article. - Tet-dependent 5-hydroxymethyl-Cytosine modification of mRNA regulates axon guidance genes in Drosophila.
Singh BN, Tran H, Kramer J, Kirichenko E, Changela N, Wang F, Feng Y, Kumar D, Tu M, Lan J, Bizet M, Fuks F, Steward R. Singh BN, et al. PLoS One. 2024 Feb 21;19(2):e0293894. doi: 10.1371/journal.pone.0293894. eCollection 2024. PLoS One. 2024. PMID: 38381741 Free PMC article.
References
- Ashburner M. 1989. Drosophila—A laboratory handbook. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Bate M. and Arias, A.M. 1993. The development of Drosophila melanogaster, pp. 149–301. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Cryderman D.E., Cuaycong, M.H., Elgin, S.C., and Wallrath, L.L. 1998. Characterization of sequences associated with position-effect variegation at pericentric sites in Drosophila heterochromatin. Chromosoma 107: 277–285. - PubMed
- Fang J., Feng, Q., Ketel, C.S., Wang, H., Cao, R., Xia, L., Erdjument-Bromage, H., Tempst, P., Simon, J.A., and Zhang, Y. 2002. Purification and functional characterization of SET8, a nucleosomal histone H4-lysine 20-specific methyltransferase. Curr. Biol. 9: 1086–1099. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials