Measuring genome conservation across taxa: divided strains and united kingdoms - PubMed (original) (raw)
Measuring genome conservation across taxa: divided strains and united kingdoms
Victor Kunin et al. Nucleic Acids Res. 2005.
Abstract
Species evolutionary relationships have traditionally been defined by sequence similarities of phylogenetic marker molecules, recently followed by whole-genome phylogenies based on gene order, average ortholog similarity or gene content. Here, we introduce genome conservation--a novel metric of evolutionary distances between species that simultaneously takes into account, both gene content and sequence similarity at the whole-genome level. Genome conservation represents a robust distance measure, as demonstrated by accurate phylogenetic reconstructions. The genome conservation matrix for all presently sequenced organisms exhibits a remarkable ability to define evolutionary relationships across all taxonomic ranges. An assessment of taxonomic ranks with genome conservation shows that certain ranks are inadequately described and raises the possibility for a more precise and quantitative taxonomy in the future. All phylogenetic reconstructions are available at the genome phylogeny server: http://maine.ebi.ac.uk:8000/cgi-bin/gps/GPS.pl.
Figures
Figure 1
Part of the complete tree of life containing the Proteobacteria generated by genome conservation (A) and gene content (B) methods. Classes are color-coded, and the Spirochaetum Leptospira interrogans and deeply branching Aquifex aeolicus are shown in black. Trees were generated using D2 normalization as described in Materials and Methods; the complete tree is available in Supplement 1.
Figure 2
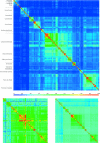
Similarity matrices across all completely sequenced organisms, derived from genome conservation (A), gene content (B) and average ortholog similarity (C). Each matrix element represents a pairwise comparison of the corresponding genomes. Genome conservation and gene content were computed using D1 normalization (see Materials and Methods). Species are ordered consistently across the different matrices, sorted according to their position on the genome conservation tree (Supplement 1), and major clades are indicated in (A). The conservation levels in percentages are color-coded, and the values for individual pairwise scores for genome conservation are available (Supplement 3). It is evident that there are three fields of values, seen as lighter blue sub-matrices representing Eukarya, Archaea and Bacteria, from top left to bottom right in (A). The diagonal values of 100% represent self-similarity. Highly similar groups are evident, for instance Escherichia coli strains (red or yellow) and enterobacteria (green), both within γ-proteobacteria. For the comparison of the matrices, see text.
Figure 3
Genome conservation within bacterial taxonomic ranks. Error bars mark standard deviations. See text for discussion, genome conservation computed using D1 normalization (see Materials and Methods).
References
- Fox G.E., Stackebrandt E., Hespell R.B., Gibson J., Maniloff J., Dyer T.A., Wolfe R.S., Balch W.E., Tanner R.S., Magrum L.J., et al. The phylogeny of prokaryotes. Science. 1980;209:457–463. - PubMed
- Doolittle R.F. Similar amino acid sequences: chance or common ancestry? Science. 1981;214:149–159. - PubMed
- Qi J., Wang B., Hao B.I. Whole proteome prokaryote phylogeny without sequence alignment: a K-string composition approach. J. Mol. Evol. 2004;58:1–11. - PubMed
- Brown J.R., Douady C.J., Italia M.J., Marshall W.E., Stanhope M.J. Universal trees based on large combined protein sequence data sets. Nature Genet. 2001;28:281–285. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources