Composition and histone substrates of polycomb repressive group complexes change during cellular differentiation - PubMed (original) (raw)
Composition and histone substrates of polycomb repressive group complexes change during cellular differentiation
Andrei Kuzmichev et al. Proc Natl Acad Sci U S A. 2005.
Abstract
Changes in the substrate specificities of factors that irreversibly modify the histone components of chromatin are expected to have a profound effect on gene expression through epigenetics. Ezh2 is a histone-lysine methyltransferase with activity dependent on its association with other components of the Polycomb Repressive Complexes 2 and 3 (PRC2/3). Ezh2 levels are increasingly elevated during prostate cancer progression. Other PRC2/3 components also are elevated in cancer cells. Overexpression of Ezh2 in tissue culture promotes formation of a previously undescribed PRC complex, PRC4, that contains the NAD+-dependent histone deacetylase SirT1 and isoform 2 of the PRC component Eed. Eed2 is expressed in cancer and undifferentiated embryonic stem (ES) cells but is undetectable in normal and differentiated ES cells. The distinct PRCs exhibit differential histone substrate specificities. These findings suggest that formation of a transformation-specific PRC complex may have a major role in resetting patterns of gene expression by regulating chromatin structure.
Figures
Fig. 1.
Characterization of histone H1 methyltransferase activity and PRC4 substrate specificity. (A) Histone H1 methylation assay of the first purification step, DE-52 column, showing separation of two histone H1-specific activities, H1KMT-1 and H1KMT-2. The assay was performed using H1-containing oligonucleosomes. Positions of H1 and core histones are indicated. Fraction numbers are indicated at the top. The fractions corresponding to H1KMT-1 activity were pooled as indicated. (B) Gel filtration analysis of the PRC4 complex. Fraction numbers and corresponding molecular mass standards are indicated at the top. The first five rows from the top are Western blots of column fractions using the indicated antibodies. The bottom three rows are HKMT assays of column fractions performed with substrates indicated on the left. Positions of core histones and histone H1 are indicated. A trace amount of nucleosomal H4 specific activity was detected in the fractions eluting at ≈1.5 MDa, likely due to a contaminating HMT activity. (C) H1 and native nucleosomes were assayed in the presence or absence of native PRC2. (D) PRC4 histone methyltransferase activity toward H1 in the presence and absence of native or recombinant oligonucleosomes. (E) Comparison of PRC2 and PRC4 methylation of H1 isoforms. Recombinant H1b, H1d, or H1o were purified from Escherichia coli and used as substrate with native oligonucleosomes. (F) PRC4 methylated K26 residue on H1b. Recombinant H1b WT or mutant with a substitution of K26 to A were used as substrates for methylation by PRC4. On the right is a comparison of the amino acid sequences of H1b, H1d, or H1o sequences with the K26 residue underlined.
Fig. 2.
Interaction between SirT1 and PRC components. (A) RNA interference (RNAi) experiments using SMARTpool of dsRNA for SirT1 (Dharmacon, Lafayette, CO) as described in ref. . HeLa cells were transfected with or without the SirT1 SMARTpool and analyzed by Western blot for the presence of SirT1, actin, H1, and H1-AcK26. (B) We transfected 293 cells with expression vectors as indicated. Extracts from transfected cells were immunoprecipitated with anti-FLAG antibodies. Aliquots of the immunoprecipitation input (in), flowthrough (ft), and eluate (ip) were analyzed by Western blot using the indicated antibodies. (C) Analysis of specificity of SirT1-Ezh2 interaction. Anti-FLAG immunoprecipitation was performed with extracts from 293 cells transfected with expression vectors encoding untagged Ezh2 and FLAG-tagged SirT proteins as follows: WT SirT1 (SirT1), active site mutant SirT1 (SirT1m), or WT SirT2 protein (SirT2). Inputs for immunoprecipitation (Input, Left) and anti-FLAG immunoprecipitates (α-FLAG ip, Right) were analyzed by Western blot using the indicated antibodies. (D) Recombinant Ezh2, Su(z)12, EED (30–535), or EED (95–535) were incubated separately with or without recombinant SirT1 protein and immunoprecipitated by using SirT1-specific antibody (2G10) coupled to beads. Inputs and elutions were analyzed by Western blot using the indicated antibodies. (E) Chromatin immunoprecipitation experiments by using an antibody to Gal4 were performed as described in ref. in 293f cells expressing Gal4Sirt1 in the presence(+Tet) or absence (–Tet) of tetracycline. The precipitated chromatin was analyzed with PCR by using primers specific to the promoters targeted by PRC2/3 (MYT1, CNR1, KCNA1, and WNT1) and to the negative control promoters (DHFR and PLCB4) as indicated. The relative enrichment of the signal in the absence or presence of tetracycline (which causes the induction of Gal4-SirT1) is shown.
Fig. 3.
Expression of PRC components is modulated as a function of differentiation and cellular transformation. (A) Western blot analysis of nuclear extracts from PGK12.1 ES cells and differentiated cells. Days of differentiation are indicated. The Western blot was probed with antibodies against SirT1, Ezh2, and Eed as well as control antibody directed against histone H3. (B) Western blot analysis of PRC components using HeLa cell nuclear extracts or whole-tissue extracts prepared from normal (N) and tumor (T) tissues obtained from a breast cancer patient and a colon cancer patient. Tissue samples obtained from the Cooperative Human Tissue Network were processed as described in ref. . The blots were probed with antibodies to Su(z)12, SirT1, Eed, and actin, which served as a loading control. A long exposure of the same blot containing the normal colon extract probed with Eed antibody is also shown.
Fig. 4.
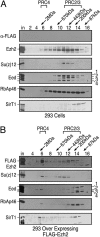
PRC complexes in 293f cells overexpressing Ezh2. Gel filtration of nuclear extracts from 293f (A) and 293-FLAG-Ezh2 (B) cell lines on Sephacryl-400 columns. Fraction numbers and molecular mass standards are indicated; “in” corresponds to column input. Western blots were probed with antibodies against proteins as indicated. The amount of protein analyzed in A was approximately twice that of B.
Fig. 5.
PRC4 analyses in vivo. (a) Immunohistochemical detection of SirT1 and Ezh2 in PIN and cancer lesions. Sections from anterior prostates of WT and Nkx3.1+/–; Pten+/– mice were processed for hematoxylin/eosin (H&E) staining (A_–_C) or immunostained by using Ezh2 (D_–_F), SirT1 (G_–_I), or Ki67 (J_–_L) antisera. (A, D, G, and J) Sections from a WT Nkx3.1+/+; Pten+/+ mouse at 9 months of age. (B, E, H, and K) Sections from a Nkx3.1+_/–; Pten+/– mouse at 12 months of age. (C, F, I, and L) Sections from a Nkx3.1+/–; Pten+/_– mouse at 15 months of age. Note that Ezh2 immunostaining is infrequent in normal tissue but is more common in PIN and carcinoma, whereas SirT1 is broadly expressed in normal epithelium and stroma but is up-regulated in epithelial cells in PIN and carcinoma. (b and c) Expression profiling of PRC components and target genes from normal to cancer lesions of prostate from compound mutant mice performed by using RNA obtained by laser-capture microdissection. Tree (b) and quantitative (c) representations of microarray results.
Similar articles
- Expression changes in EZH2, but not in BMI-1, SIRT1, DNMT1 or DNMT3B are associated with DNA methylation changes in prostate cancer.
Hoffmann MJ, Engers R, Florl AR, Otte AP, Muller M, Schulz WA. Hoffmann MJ, et al. Cancer Biol Ther. 2007 Sep;6(9):1403-12. doi: 10.4161/cbt.6.9.4542. Cancer Biol Ther. 2007. PMID: 18637271 - The polycomb group gene product Ezh2 regulates proliferation and differentiation of murine hepatic stem/progenitor cells.
Aoki R, Chiba T, Miyagi S, Negishi M, Konuma T, Taniguchi H, Ogawa M, Yokosuka O, Iwama A. Aoki R, et al. J Hepatol. 2010 Jun;52(6):854-63. doi: 10.1016/j.jhep.2010.01.027. Epub 2010 Mar 24. J Hepatol. 2010. PMID: 20395008 - Importance of Ezh2 polycomb protein in tumorigenesis process interfering with the pathway of growth suppressive key elements.
Tonini T, D'Andrilli G, Fucito A, Gaspa L, Bagella L. Tonini T, et al. J Cell Physiol. 2008 Feb;214(2):295-300. doi: 10.1002/jcp.21241. J Cell Physiol. 2008. PMID: 17786943 Review. - The EZH2 polycomb transcriptional repressor--a marker or mover of metastatic prostate cancer?
Sellers WR, Loda M. Sellers WR, et al. Cancer Cell. 2002 Nov;2(5):349-50. doi: 10.1016/s1535-6108(02)00187-3. Cancer Cell. 2002. PMID: 12450788 Review.
Cited by
- Epigenome remodelling in breast cancer: insights from an early in vitro model of carcinogenesis.
Locke WJ, Clark SJ. Locke WJ, et al. Breast Cancer Res. 2012 Nov 15;14(6):215. doi: 10.1186/bcr3237. Breast Cancer Res. 2012. PMID: 23168266 Free PMC article. Review. - Aberrant cytoplasm localization and protein stability of SIRT1 is regulated by PI3K/IGF-1R signaling in human cancer cells.
Byles V, Chmilewski LK, Wang J, Zhu L, Forman LW, Faller DV, Dai Y. Byles V, et al. Int J Biol Sci. 2010 Oct 7;6(6):599-612. doi: 10.7150/ijbs.6.599. Int J Biol Sci. 2010. PMID: 20941378 Free PMC article. - Inhibition of SIRT1 reactivates silenced cancer genes without loss of promoter DNA hypermethylation.
Pruitt K, Zinn RL, Ohm JE, McGarvey KM, Kang SH, Watkins DN, Herman JG, Baylin SB. Pruitt K, et al. PLoS Genet. 2006 Mar;2(3):e40. doi: 10.1371/journal.pgen.0020040. Epub 2006 Mar 31. PLoS Genet. 2006. PMID: 16596166 Free PMC article. - Control of developmental regulators by Polycomb in human embryonic stem cells.
Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, Kumar RM, Chevalier B, Johnstone SE, Cole MF, Isono K, Koseki H, Fuchikami T, Abe K, Murray HL, Zucker JP, Yuan B, Bell GW, Herbolsheimer E, Hannett NM, Sun K, Odom DT, Otte AP, Volkert TL, Bartel DP, Melton DA, Gifford DK, Jaenisch R, Young RA. Lee TI, et al. Cell. 2006 Apr 21;125(2):301-13. doi: 10.1016/j.cell.2006.02.043. Cell. 2006. PMID: 16630818 Free PMC article. - Epigenetic remodeling of chromatin architecture: exploring tumor differentiation therapies in mesenchymal stem cells and sarcomas.
Siddiqi S, Mills J, Matushansky I. Siddiqi S, et al. Curr Stem Cell Res Ther. 2010 Mar;5(1):63-73. doi: 10.2174/157488810790442859. Curr Stem Cell Res Ther. 2010. PMID: 19807660 Free PMC article. Review.
References
- Jenuwein, T. & Allis, C. D. (2001) Science 293, 1074–1080. - PubMed
- Sims, R. J., III, Nishioka, K. & Reinberg, D. (2003) Trends Genet. 19, 629–639. - PubMed
- Lachner, M. & Jenuwein, T. (2002) Curr. Opin. Cell Biol. 14, 286–298. - PubMed
- Cao, R., Wang, L., Wang, H., Xia, L., Erdjument-Bromage, H., Tempst, P., Jones, R. S. & Zhang, Y. (2002) Science 298, 1039–1043. - PubMed
- Czermin, B., Melfi, R., McCabe, D., Seitz, V., Imhof, A. & Pirrotta, V. (2002) Cell 111, 185–196. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases