Molecular characterization of proteolytic cleavage sites of the Pseudomonas syringae effector AvrRpt2 - PubMed (original) (raw)
Molecular characterization of proteolytic cleavage sites of the Pseudomonas syringae effector AvrRpt2
Stephen T Chisholm et al. Proc Natl Acad Sci U S A. 2005.
Abstract
During infection of Arabidopsis thaliana, the bacterium Pseudomonas syringae pv tomato delivers the effector protein AvrRpt2 into the plant cell cytosol. Within the plant cell, AvrRpt2 undergoes N-terminal processing and causes elimination of Arabidopsis RIN4. Previous work established that AvrRpt2 is a putative cysteine protease, and AvrRpt2 processing and RIN4 elimination require an intact predicted catalytic triad in that AvrRpt2. In this work, proteolytic events that depend on AvrRpt2 activity were characterized. The amino acid sequence surrounding the processing site of AvrRpt2 and two related sequences from RIN4 triggered Avr-Rpt2-dependent proteolytic cleavage of a synthetic substrate, demonstrating that these sequences are cleavage recognition sites for AvrRpt2 activity. Processing-deficient AvrRpt2 mutants were identified and shown to retain their ability to eliminate wild-type RIN4. Single amino acid substitutions were made in each of the two RIN4 cleavage sites, and mutation of both sites resulted in cleavage-resistant RIN4. Growth of Pseudomonas expressing AvrRpt2 was significantly higher than catalytically inactive mutants on Arabidopsis rin4/rps2 mutant plants, suggesting there are additional protein targets of AvrRpt2 that account for the virulence activity of this effector. Bioinformatics analysis identified putative Arabidopsis proteins containing sequences similar to the proteolytic cleavage sites conserved in AvrRpt2 and RIN4. Several of these proteins were eliminated in an AvrRpt2-dependent manner in a transient in planta expression system. These results identify amino acids important for AvrRpt2 substrate recognition and cleavage as well as demonstrate AvrRpt2 protease activity eliminates multiple Arabidopsis proteins in a transient expression system.
Figures
Fig. 1.
AvrRpt2-dependent cleavage of synthetic substrates. (A) Alignment of the amino acid sequence surrounding the cleavage site (filled arrowhead) of AvrRpt2 (ACS) with sequences from RIN4 (RCS1 and RCS2). Positions of these amino acids in the respective full-length proteins are given in the left column. Amino acids conserved in all sites are boxed. (B) Schematic representation of synthetic substrate. A translational fusion was generated between GFP and GUS such that the sequences indicated in A could be cloned between GFP and GUS. Single-letter codes are given for amino acids encoded by _Sal_I, _Xma_I, and _Sac_I restriction sites used for cloning purposes. (C) AvrRpt2-dependent cleavage of synthetic substrates. Immunoblots of protein extracts from N. benthamiana leaves transiently expressing synthetic substrates containing the indicated putative cleavage sites alone (lane –), with AvrRpt2:FLAG (lane A) or AvrRpt2:FLAG(C122A) (lane C). Resolved extracts were probed with αGUS (Upper) and αGFP (Lower). A crossreacting protein is indicated (shaded circle). Approximate positions of molecular mass standards are shown in kilodaltons.
Fig. 2.
Processing of AvrRpt2:HA mutants in rabbit reticulocyte lysates. The indicated forms of AvrRpt2:HA were expressed from a T7 promoter in rabbit reticulocyte lysates in the presence of [35S]methionine. After the addition of template DNA, protein samples were collected at the indicated time points and resolved by using denaturing PAGE. A phosphorimaging screen was used to detect radiolabeled proteins. (Upper) Control reactions. (Lower) Conserved residues (numbered, filled amino acids) of the AvrRpt2:HA cleavage site were substituted with the indicated amino acids and analyzed as above. Filled arrowheads, unprocessed AvrRpt2:HA; open arrowheads, processed AvrRpt2:HA; filled circles, background protein bands.
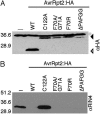
Fig. 3.
Processing-deficient AvrRpt2:HA mutants prevent T7:RIN4 accumulation in planta. The indicated forms of AvrRpt2:HA (–, no AvrRpt2:HA) were coexpressed with T7:RIN4 in N. benthamiana leaves. Protein extracts were resolved by denaturing PAGE and used for immunoblot analyses. Approximate positions of molecular mass standards are shown. (A) Processing of AvrRpt2:HA forms in planta. AvrRpt2:HA (WT) accumulates as processed protein (open arrowhead). Catalytically deficient AvrRpt2:HA(C122A) and ACS mutants AvrRpt2:HA(F70A/G71A), (F70R), and (ΔPAFGG) accumulate as unprocessed proteins (filled arrowhead). Filled circle, crossreacting protein. (B) T7:RIN4 does not accumulate in the presence of processing-deficient AvrRpt2:HA. RIN4 antisera detects T7:RIN4 expressed alone (–) or with AvrRpt2:HA(C122A). T7:RIN4 is not detected when coexpressed with AvrRpt2:HA or AvrRpt2:HA(F70A/G71A), (F70R), or (ΔPAFGG).
Fig. 4.
T7:RIN4 is cleaved at two sites in planta.(A) T7:RIN4 or the indicated T7:RIN4 mutants were coexpressed in N. benthamiana leaves with RPS2:HA and GFP (lane g), AvrRpt2:FLAG (lane A), or AvrRpt2:FLAG(C122A) (lane C). X, noninfiltrated leaf. Protein extracts were resolved by denaturing PAGE and used for immunoblot analyses with αRIN4. Approximate positions of molecular mass standards are shown. (B) RIN4 amino acid sequence. Boxed regions, conserved portions of AvrRpt2 cleavage recognition sequences; underlined regions, additional occurrences of FG sequence.
Fig. 5.
AvrRpt2-dependent cleavage of Arabidopsis proteins. (A) AvrRpt2 contributes to bacterial virulence on Arabidopsis rin4/rps2 plants. P. syringae pv tomato strain DC3000 (Pst) expressing GFP, AvrRpt2, or catalytically deficient AvrRpt2 C122A or H208A were inoculated into Arabidopsis Col-0 and the indicated Col-0 mutant lines. Bacterial growth was quantified at 4 days postinoculation. (B) Alignment of putative ACS in Arabidopsis proteins. The Arabidopsis locus identifier is shown, followed by the start index, the sequence, and the end index of the site. RCS1 and RCS2 are represented as RIN4_1 and RIN4_2. Completely conserved residues are black, and similar residues are gray. (C) Putative targets do not accumulate in the presence of AvrRpt2. The indicated epitope-tagged proteins were expressed in N. benthamiana leaves alone (–), with AvrRpt2:FLAG (lane A), or with AvrRpt2:FLAG(C122A) (lane C). X, noninfiltrated leaf. Putative cleavage site amino acid sequences are given in parentheses. Protein extracts were resolved by denaturing PAGE and used for immunoblot analyses with αT7. Approximate positions of molecular mass standards are shown. (D) Mutation of putative cleavage sites prevents AvrRpt2-dependent protein elimination. The indicated epitope-tagged proteins were expressed in N. benthamiana and analyzed as in C. Amino acid mutations for each protein are listed in parentheses.
Similar articles
- The Pseudomonas syringae effector AvrRpt2 cleaves its C-terminally acylated target, RIN4, from Arabidopsis membranes to block RPM1 activation.
Kim HS, Desveaux D, Singer AU, Patel P, Sondek J, Dangl JL. Kim HS, et al. Proc Natl Acad Sci U S A. 2005 May 3;102(18):6496-501. doi: 10.1073/pnas.0500792102. Epub 2005 Apr 21. Proc Natl Acad Sci U S A. 2005. PMID: 15845764 Free PMC article. - Genetic and molecular evidence that the Pseudomonas syringae type III effector protein AvrRpt2 is a cysteine protease.
Axtell MJ, Chisholm ST, Dahlbeck D, Staskawicz BJ. Axtell MJ, et al. Mol Microbiol. 2003 Sep;49(6):1537-46. doi: 10.1046/j.1365-2958.2003.03666.x. Mol Microbiol. 2003. PMID: 12950919 - Role of RIN4 in Regulating PAMP-Triggered Immunity and Effector-Triggered Immunity: Current Status and Future Perspectives.
Ray SK, Macoy DM, Kim WY, Lee SY, Kim MG. Ray SK, et al. Mol Cells. 2019 Jul 31;42(7):503-511. doi: 10.14348/molcells.2019.2433. Mol Cells. 2019. PMID: 31362467 Free PMC article. Review. - Plant defense: one post, multiple guards?!
Marathe R, Dinesh-Kumar SP. Marathe R, et al. Mol Cell. 2003 Feb;11(2):284-6. doi: 10.1016/s1097-2765(03)00072-8. Mol Cell. 2003. PMID: 12620215 Review.
Cited by
- Phosphorylation of the Plant Immune Regulator RPM1-INTERACTING PROTEIN4 Enhances Plant Plasma Membrane H⁺-ATPase Activity and Inhibits Flagellin-Triggered Immune Responses in Arabidopsis.
Lee D, Bourdais G, Yu G, Robatzek S, Coaker G. Lee D, et al. Plant Cell. 2015 Jul;27(7):2042-56. doi: 10.1105/tpc.114.132308. Epub 2015 Jul 21. Plant Cell. 2015. PMID: 26198070 Free PMC article. - Activation of a plant nucleotide binding-leucine rich repeat disease resistance protein by a modified self protein.
DeYoung BJ, Qi D, Kim SH, Burke TP, Innes RW. DeYoung BJ, et al. Cell Microbiol. 2012 Jul;14(7):1071-84. doi: 10.1111/j.1462-5822.2012.01779.x. Epub 2012 Mar 27. Cell Microbiol. 2012. PMID: 22372664 Free PMC article. - AvrRpm1 missense mutations weakly activate RPS2-mediated immune response in Arabidopsis thaliana.
Cherkis KA, Temple BR, Chung EH, Sondek J, Dangl JL. Cherkis KA, et al. PLoS One. 2012;7(8):e42633. doi: 10.1371/journal.pone.0042633. Epub 2012 Aug 6. PLoS One. 2012. PMID: 22880057 Free PMC article. - The Pseudomonas syringae effector AvrRpt2 cleaves its C-terminally acylated target, RIN4, from Arabidopsis membranes to block RPM1 activation.
Kim HS, Desveaux D, Singer AU, Patel P, Sondek J, Dangl JL. Kim HS, et al. Proc Natl Acad Sci U S A. 2005 May 3;102(18):6496-501. doi: 10.1073/pnas.0500792102. Epub 2005 Apr 21. Proc Natl Acad Sci U S A. 2005. PMID: 15845764 Free PMC article. - Type III effector activation via nucleotide binding, phosphorylation, and host target interaction.
Desveaux D, Singer AU, Wu AJ, McNulty BC, Musselwhite L, Nimchuk Z, Sondek J, Dangl JL. Desveaux D, et al. PLoS Pathog. 2007 Mar;3(3):e48. doi: 10.1371/journal.ppat.0030048. PLoS Pathog. 2007. PMID: 17397263 Free PMC article.
References
- Galan, J. E. & Collmer, A. (1999) Science 284, 1322–1328. - PubMed
- Alfano, J. R. & Collmer, A. (2004) Annu. Rev. Phytopathol. 42, 385–414. - PubMed
- Chen, Z., Kloek, A. P., Boch, J., Katagiri, F. & Kunkel, B. N. (2000) Mol. Plant–Microbe Interact. 13, 1312–1321. - PubMed
- Abramovitch, R. B. & Martin, G. B. (2004) Curr. Opin. Plant Biol. 7, 356–364. - PubMed
- Nimchuk, Z., Eulgem, T., Holt, B. F., III, & Dangl, J. L. (2003) Annu. Rev. Genet. 37, 579–609. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous