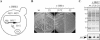Partial depletion of histone H4 increases homologous recombination-mediated genetic instability - PubMed (original) (raw)
Partial depletion of histone H4 increases homologous recombination-mediated genetic instability
Félix Prado et al. Mol Cell Biol. 2005 Feb.
Abstract
DNA replication can be a source of genetic instability. Given the tight connection between DNA replication and nucleosome assembly, we analyzed the effect of a partial depletion of histone H4 on genetic instability mediated by homologous recombination. A Saccharomyces cerevisiae strain was constructed in which the expression of histone H4 was driven by the regulated tet promoter. In agreement with defective nucleosome assembly, partial depletion of histone H4 led to subtle changes in plasmid superhelical density and chromatin sensitivity to micrococcal nuclease. Under these conditions, homologous recombination between ectopic DNA sequences was increased 20-fold above the wild-type levels. This hyperrecombination was not associated with either defective repair or transcription but with an accumulation of recombinogenic DNA lesions during the S and G(2)/M phases, as determined by an increase in the proportion of budded cells containing Rad52-yellow fluorescent protein foci. Consistently, partial depletion of histone H4 caused a delay during the S and G(2)/M phases. Our results suggest that histone deposition defects lead to the formation of recombinogenic DNA structures during replication that increase genomic instability.
Figures
FIG. 1.
Construction of a yeast strain (t::HHF2) that can be gradually depleted of histone H4. (A) Scheme of the main genotype of strain t::HHF2. This strain is a null mutant for the two copies of the histone H4 gene (_hhf1_Δ _hhf2_Δ) that contains a plasmid (p413TARtetH4) expressing HHF2 under the control of the bacterial tet promoter, which responds to DOX in a dose-dependent manner. (B) Growth analysis of BYtetH4-10D at 5 (t::HHF2[5]) or 0.25 (t::HHF2[0.25]) μg of DOX/ml or in the absence of DOX and of BY4741 at 5 μg of DOX/ml (wild type [wt]). Cell growth of BY4741 was not affected by the presence or absence of DOX (data not shown). (C) Histone H4 content from strains described in panel B as determined by Western blot analysis. Total protein content was determined by Coomassie staining. The luminescence signals corresponding to histone H4 in t::HHF2[5] and t::HHF2[0.25] cells were 71 and 52% of the wild-type levels, respectively.
FIG. 2.
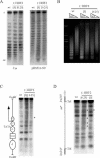
Analysis of plasmid superhelical density, chromatin accessibility, and nucleosome positioning in wild-type and t::HHF2 cells. (A) Topoisomer distribution of centromeric pRS316-SU and multicopy 2μm plasmids after electrophoresis in agarose gels containing 4 μg of chloroquine/ml. At this chloroquine concentration, negatively supercoiled topoisomers are resolved, consistent with a slower migration with 8 μg of chloroquine/ml (data not shown). r and sc indicate relaxed and negatively supercoiled plasmids, respectively. (B) MNase accessibility of bulk chromatin. MN, MNase I. (C and D) MNase digestion patterns of the endogenous GAL1 gene and the intervening sequence between the inverted leu2 repeats of the SU repeat system, respectively. A scheme of the analyzed sequences is shown on the left of each panel. Bands whose intensity is modified relative to the wild type are indicated with asterisks. Strains and other details are the same as for Fig. 1B.
FIG. 3.
Recombination in cells partially depleted of histone H4. (A) Frequencies of Leu+ recombinants in BY4741 (wild type [wt]), Y03144 (_hhf1_Δ), and Y15356 (_hhf2_Δ) at 5 μg of DOX/ml and BYtetH4-10D at either 5 (t::HHF2[5]) or 0.25 (t::HHF2[0.25]) μg of DOX/ml. (B) Frequencies of Leu+ recombinants in BY4741 (wt), BY51 (_rad51_Δ), BY59 (_rad59_Δ), Y10540 (_rad52_Δ), BYtetH4-10D (t::HHF2[5]), BY51tetH4 (t::HHF2[5] _rad51_Δ), BY59tetH4 (t::HHF2[5] _rad59_Δ), and BY52tetH4 (t::HHF2[5] _rad52_Δ) at 5 μg of DOX/ml. All strains carried either the pRS316-LYΔNS or the pRS316-SU plasmid, harboring the direct repeat system LYΔNS or the inverted repeat system SU, respectively. A scheme of each recombination system is shown at the top of the panels. The frequencies of Leu+ recombinants of wild-type, _hhf1_Δ, _hhf2_Δ, _rad51_Δ, _rad52_Δ, and _rad59_Δ strains were not affected by the presence or absence of DOX (data not shown). The averages and standard deviations of three or four median frequencies obtained with two or three independent transformants are shown.
FIG. 4.
DNA repair in cells partially depleted of histone H4. (A) Growth analysis of BYtetH4-10D (t::HHF2) and BY52tetH4 (t::HHF2 _rad52_Δ) at either 5 (t::HHF2[5]) or 0.25 (t::HHF2[0.25]) μg of DOX/ml. (B, C, and D) UV light, HU, and MMS sensitivities of the strains indicated in the legend to Fig. 1, respectively. _rad1_Δ, _asf1_Δ, and _rad52_Δ strains are controls for UV light-, HU-, and MMS-induced DNA repair, respectively, and were analyzed at both 5 (t::HHF2[5]) and 0.25 (t::HHF2[0.25]) μg of DOX/ml. Since UV light, HU, and MMS sensitivities of wild-type, _rad52_Δ, _asf1_Δ, and _rad1_Δ strains were not affected by the presence or absence of DOX (data not shown), only the data for 5 μg of DOX/ml are shown for these strains.
FIG. 5.
Effect of transcription in recombination induced by partial depletion of histone H4. (A) leu2 RNA levels and frequencies of Leu+ recombinants of strains Y07202 (wild type [wt]) and BYtetH4-10D (t::HHF2[5]) transformed with pRS314-LNA or pRS314-LNAT and grown in 5 μg of DOX/ml. (B) leu2 RNA levels and frequencies of Leu+ recombinants of strains Y07202 (wt) and BYth2-1B (_hhf2_Δ) transformed with pRS314-GL and grown in either glucose- or galactose-containing medium, in which the GAL1 promoter of the GL system is either repressed or activated, respectively. Schemes of the direct repeat systems are shown. Dashed arrows indicate the transcripts produced by the recombination systems. As a probe, the ClaI-EcoRV LEU2 fragment was used. The averages and standard deviations of two to four median frequencies obtained with two to four independent transformants are shown.
FIG. 6.
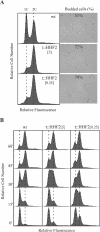
Cell cycle analysis of cells partially depleted of histone H4. (A) DNA content and cell morphology of asynchronous cultures of the strains and at the conditions indicated in the legend to Fig. 1. Similar profiles were obtained with BYtetH4l-1C (data not shown). (B) Cell cycle progression of BYtetH4l-1C at either 5 (t::HHF2[5]) or 0.25 (t::HHF2[0.25]) μg of DOX/ml and of BY4741 at 5 μg of DOX/ml (wild type [wt]). DNA content of wild-type and t::HHF2 cells was determined by flow cytometry at the indicated times upon release from α-factor arrest in G1.
FIG. 7.
(A) Effect of HU on recombination induced by partial depletion of histone H4. Frequencies of Leu+ recombinants of the strains indicated in the legend to Fig. 1 grown with 5 μg of DOX/ml and either 0, 25, or 100 mM HU. (B) Accumulation of Rad52-YFP foci in t::HHF2 cells. Rad52-YFP foci were visualized by fluorescence microscopy in asynchronous cultures of BYtetH4-10D transformed with pWJ1344 and grown in either 5 (t::HHF2[5]) or 0.25 (t::HHF2[0.25]) μg of DOX/ml and of BY4741 transformed with pWJ1344 and grown in 5 μg of DOX/ml (wild type [wt]). The percentages of unbudded (G1) and budded (S and G2/M) wild-type and t::HHF2 cells containing Rad52-YFP foci are shown. The total numbers of analyzed cells were 50 for unbudded cells and 150 for budded cells. The averages and standard deviations of two independent experiments are plotted.
Similar articles
- Evidence for distinct mechanisms facilitating transcript elongation through chromatin in vivo.
Kristjuhan A, Svejstrup JQ. Kristjuhan A, et al. EMBO J. 2004 Oct 27;23(21):4243-52. doi: 10.1038/sj.emboj.7600433. Epub 2004 Sep 30. EMBO J. 2004. PMID: 15457216 Free PMC article. - Regulation of NuA4 histone acetyltransferase activity in transcription and DNA repair by phosphorylation of histone H4.
Utley RT, Lacoste N, Jobin-Robitaille O, Allard S, Côté J. Utley RT, et al. Mol Cell Biol. 2005 Sep;25(18):8179-90. doi: 10.1128/MCB.25.18.8179-8190.2005. Mol Cell Biol. 2005. PMID: 16135807 Free PMC article. - H4 acetylation does not replace H3 acetylation in chromatin remodelling and transcription activation of Adr1-dependent genes.
Agricola E, Verdone L, Di Mauro E, Caserta M. Agricola E, et al. Mol Microbiol. 2006 Dec;62(5):1433-46. doi: 10.1111/j.1365-2958.2006.05451.x. Mol Microbiol. 2006. PMID: 17121596 - Genome-wide patterns of histone modifications in yeast.
Millar CB, Grunstein M. Millar CB, et al. Nat Rev Mol Cell Biol. 2006 Sep;7(9):657-66. doi: 10.1038/nrm1986. Epub 2006 Aug 16. Nat Rev Mol Cell Biol. 2006. PMID: 16912715 Review. - Mitotic recombination in yeast: elements controlling its incidence.
Aguilera A, Chávez S, Malagón F. Aguilera A, et al. Yeast. 2000 Jun 15;16(8):731-54. doi: 10.1002/1097-0061(20000615)16:8<731::AID-YEA586>3.0.CO;2-L. Yeast. 2000. PMID: 10861900 Review.
Cited by
- The histone H2B-specific ubiquitin ligase RNF20/hBRE1 acts as a putative tumor suppressor through selective regulation of gene expression.
Shema E, Tirosh I, Aylon Y, Huang J, Ye C, Moskovits N, Raver-Shapira N, Minsky N, Pirngruber J, Tarcic G, Hublarova P, Moyal L, Gana-Weisz M, Shiloh Y, Yarden Y, Johnsen SA, Vojtesek B, Berger SL, Oren M. Shema E, et al. Genes Dev. 2008 Oct 1;22(19):2664-76. doi: 10.1101/gad.1703008. Genes Dev. 2008. PMID: 18832071 Free PMC article. - Highly conserved regimes of neighbor-base-dependent mutation generated the background primary-structural heterogeneities along vertebrate chromosomes.
Antezana MA, Jordan IK. Antezana MA, et al. PLoS One. 2008 May 14;3(5):e2145. doi: 10.1371/journal.pone.0002145. PLoS One. 2008. PMID: 18478116 Free PMC article. - Defective histone supply causes condensin-dependent chromatin alterations, SAC activation and chromosome decatenation impairment.
Murillo-Pineda M, Cabello-Lobato MJ, Clemente-Ruiz M, Monje-Casas F, Prado F. Murillo-Pineda M, et al. Nucleic Acids Res. 2014 Nov 10;42(20):12469-82. doi: 10.1093/nar/gku927. Epub 2014 Oct 9. Nucleic Acids Res. 2014. PMID: 25300489 Free PMC article. - New histone supply regulates replication fork speed and PCNA unloading.
Mejlvang J, Feng Y, Alabert C, Neelsen KJ, Jasencakova Z, Zhao X, Lees M, Sandelin A, Pasero P, Lopes M, Groth A. Mejlvang J, et al. J Cell Biol. 2014 Jan 6;204(1):29-43. doi: 10.1083/jcb.201305017. Epub 2013 Dec 30. J Cell Biol. 2014. PMID: 24379417 Free PMC article. - Regulation of Chromatin Assembly and Cell Transformation by Formaldehyde Exposure in Human Cells.
Chen D, Fang L, Mei S, Li H, Xu X, Des Marais TL, Lu K, Liu XS, Jin C. Chen D, et al. Environ Health Perspect. 2017 Sep 21;125(9):097019. doi: 10.1289/EHP1275. Environ Health Perspect. 2017. PMID: 28937961 Free PMC article.
References
- Adams, C. R., and R. T. Kamakaka. 1999. Chromatin assembly: biochemical identities and genetic redundancy. Curr. Opin. Genet. Dev. 9:185-190. - PubMed
- Adkins, M. W., S. R. Howar, and J. K. Tyler. 2004. Chromatin disassembly mediated by the histone chaperone Asf1 is essential for transcriptional activation of the yeast PHO5 and PHO8 genes. Mol. Cell 14:657-666. - PubMed
- Adkins, M. W., and J. K. Tyler. 2004. The histone chaperone Asf1p mediates chromatin disassembly in vivo. J. Biol. Chem. 279:52069-52074. - PubMed
- Aguilera, A. 1995. Genetic evidence for different RAD52-dependent intrachromosomal recombination pathways in Saccharomyces cerevisiae. Curr. Genet. 27:298-305. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials