DNA binding: a novel function of Pseudomonas aeruginosa type IV pili - PubMed (original) (raw)
DNA binding: a novel function of Pseudomonas aeruginosa type IV pili
Erin J van Schaik et al. J Bacteriol. 2005 Feb.
Abstract
The opportunistic pathogen Pseudomonas aeruginosa produces multifunctional, polar, filamentous appendages termed type IV pili. Type IV pili are involved in colonization during infection, twitching motility, biofilm formation, bacteriophage infection, and natural transformation. Electrostatic surface analysis of modeled pilus fibers generated from P. aeruginosa strain PAK, K122-4, and KB-7 pilin monomers suggested that a solvent-exposed band of positive charge may be a common feature of all type IV pili. Several functions of type IV pili, including natural transformation and biofilm formation, involve DNA. We investigated the ability of P. aeruginosa type IV pili to bind DNA. Purified PAK, K122-4, and KB-7 pili were observed to bind both bacterial plasmid and salmon sperm DNA in a concentration-dependent and saturable manner. PAK pili had the highest affinity for DNA, followed by K122-4 and KB-7 pili. DNA binding involved backbone interactions and preferential binding to pyrimidine residues even though there was no evidence of sequence-specific binding. Pilus-mediated DNA binding was a function of the intact pilus and thus required elements present in the quaternary structure. However, binding also involved the pilus tip as tip-specific, but not base-specific, antibodies inhibited DNA binding. The conservation of a Thr residue in all type IV pilin monomers examined to date, along with the electrostatic data, implies that DNA binding is a conserved function of type IV pili. Pilus-mediated DNA binding could be important for biofilm formation both in vivo during an infection and ex vivo on abiotic surfaces.
Figures
FIG. 1.
Amino acid sequence and structural aspects of P. aeruginosa type IV pilins. (A) Electrostatic surface representation generated by using Delphi in InsightII of modeled pilins assembled into a pilus fiber (as described in reference 27) from P. aeruginosa strains K122-4, PAK, and KB-7. Blue represents a positive charge, red represents a negative charge, and white represents a neutral charge. (B) Multiple-sequence alignment of type IV pilins. Strictly, highly, and moderately conserved positions are indicated by asterisks, colons, and periods, respectively, above the sequences, and conserved secondary structural elements, based on an average for solved pilin structures, are indicated below the sequences. The N-terminal α-helix is indicated by a cylinder, and β-sheets are indicated by arrows. Production of soluble pilin monomers for structural studies (2, 24, 27) is achieved by truncation of the 28 N-terminal residues, indicated by a vertical arrow. The disulfide-bound receptor binding loop is enclosed in a box. The sequences are the sequences of N. gonorrhoeae strain MS11 (gi 3212472), N. meningitidis strain FAM18 (gi 2228578), and P. aeruginosa strains K122-4 (gi 77636), PAK (gi 120438), PAO (gi 120440), KB-7 (gi 3219798), and 1244 (gi 77632). The alignment was prepared with CLUSTALW (56) with minor manual editing.
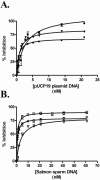
FIG. 2.
Concentration dependence of DNA binding by purified biotinylated pili from P. aeruginosa strains PAK, K122-4, and KB-7. (A and B) The pUCP19 plasmid (A) (solid symbols) or salmon sperm DNA (B) (open symbols) was immobilized in the wells of microtiter plates by using poly-
l
-lysine. Various concentrations of biotinylated PAK (▪ and □), K122-4 (• and ○), or KB-7 (▾ and ▿) pili or His-tagged PAK (♦) or K122-4 (▴) pilin were added to the plates and incubated for 1.5 h at room temperature. Binding was quantified spectrophotometrically by using streptavidin-HRP or primary mouse His antibodies and secondary HRP conjugate. The data are means and standard deviations of at least three replicates from two independent experiments. (C) ELISA of His-tagged PAK (♦) or K122-4 (▴) monomers. His-tagged monomers were immobilized in the wells of microtiter plates at a concentration of 10 μg/ml. Antibody dilutions were added to the wells containing the immobilized His-tagged monomers in the presence of 5 mM MgCl2 and incubated for 1 h at 37°C. Binding was quantified spectrophotometrically by using anti-mouse HRP. The data are the means and standard deviations of three replicates.
FIG. 3.
Exogenous DNA inhibits binding of biotinylated pili to immobilized DNA. pUCP19 plasmid DNA (A) or salmon sperm DNA (B) was immobilized in the wells of microtiter plates by using poly-
l
-lysine. Biotinylated PAK (▪ and □), K122-4 (• and ○), or KB-7 (▾ and ▿) pili were preincubated with various amounts of exogenous DNA for 30 min. The reaction mixtures were then added to microtiter plates containing immobilized DNA and incubated for 1.5 h at room temperature. Binding was quantified spectrophotometrically by using streptavidin-HRP. The data are the means and standard deviations of at least three replicates from two independent experiments.
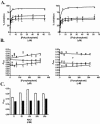
FIG. 4.
(A and B) Polyphosphate but not pyrophosphate inhibits binding of biotinylated pili to immobilized DNA. pUCP19 plasmid DNA (open symbols) or salmon sperm DNA (solid symbols) was immobilized in the wells of microtiter plates by using poly-
l
-lysine. Biotinylated PAK (▪ and □), K122-4 (• and ○), or KB-7 (▾ and ▿) pili were preincubated with various concentrations of polyphosphate (A) or pyrophosphate (B) for 30 min. The reaction mixtures were then added to microtiter plates containing immobilized DNA and incubated for 1.5 h at room temperature. Binding was quantified spectrophotometrically by using streptavidin-HRP. The data are the means and standard deviations of at least three replicates from two independent experiments. (C) Salt dependence of pilus-mediated DNA binding. Biotinylated K122-4 pili (open bars) or KB-7 pili (solid bars) were added to microtiter plates containing immobilized pUCP19 at a concentration of 0.5 μg/ml in 0.01 M PB (pH 7.4) containing various amounts of NaCl and incubated for 1.5 h at RT. Binding was quantified spectrophotometrically by using streptavidin-HRP. The data are the means and standard deviations of three replicates.
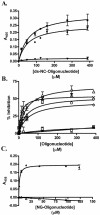
FIG. 5.
Direct and competitive binding of biotinylated oligonucleotides to immobilized P. aeruginosa pili purified from strains PAK, K122-4, and KB-7. (A) Concentration dependence of binding of a biotinylated double-stranded oligonucleotide containing the N. gonorrhoeae uptake sequence (Table 1) to immobilized PAK pili (▪), K122-4 pili (•), or KB-7 pili (▾). Various concentrations of the double-stranded NG oligonucleotide were added to the plates containing the immobilized pili and incubated for 1.5 h at room temperature. Binding was quantified spectrophotometrically by using streptavidin-HRP. The data are the means and standard deviations of at least three replicates from two independent experiments. (B) Direct competitive binding of single-stranded oligonucleotides (Table 1) and biotinylated oligonucleotide ss-NG to immobilized PAK pili. Biotinylated oligonucleotide ss-NG (15 μM) was mixed with various concentrations of unlabeled NG (▵), BLAD (×), poly(C) (○), poly(A) (□), poly(T) (⋄), or poly(G) (*) added to the microtiter plates containing immobilized PAK pili and incubated for 1.5 h. Binding was quantified spectrophotometrically by using streptavidin-HRP. The data are the means and standard deviations of at least three replicates from two independent experiments. (C) Direct binding of untreated ss-NG (▵) or S1 nuclease-treated ss-NG (▴) to immobilized PAK pili.
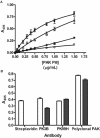
FIG. 6.
Antibody inhibition of binding of biotinylated PAK pili to immobilized pUCP19. pUCP19 was immobilized in the wells of microtiter plates by using poly-
l
-lysine. (A) Biotinylated PAK pili were either not treated (▪) or preincubated with PK99H (*), PK3B (▵), or polyclonal PAK (○) antibodies for 45 min at room temperature. The reaction mixtures were then added to microtiter plates containing immobilized plasmid pUCP19 and incubated for 1.5 h at room temperature. Binding was quantified spectrophotometrically by using streptavidin-HRP. The data are the means and standard deviations of at least three replicates from two independent experiments. (B) ELISA performed with unlabeled or biotinylated PAK pili and streptavidin-HRP, PK99H, PK3B, and polyclonal PAK. The data are the means and standard deviation of three replicate wells.
Similar articles
- The Pseudomonas aeruginosa type IV pilin receptor binding domain functions as an adhesin for both biotic and abiotic surfaces.
Giltner CL, van Schaik EJ, Audette GF, Kao D, Hodges RS, Hassett DJ, Irvin RT. Giltner CL, et al. Mol Microbiol. 2006 Feb;59(4):1083-96. doi: 10.1111/j.1365-2958.2005.05002.x. Mol Microbiol. 2006. PMID: 16430686 - Crystallographic analysis of the Pseudomonas aeruginosa strain K122-4 monomeric pilin reveals a conserved receptor-binding architecture.
Audette GF, Irvin RT, Hazes B. Audette GF, et al. Biochemistry. 2004 Sep 14;43(36):11427-35. doi: 10.1021/bi048957s. Biochemistry. 2004. PMID: 15350129 - Pseudomonas aeruginosa Type IV pilus expression in Neisseria gonorrhoeae: effects of pilin subunit composition on function and organelle dynamics.
Winther-Larsen HC, Wolfgang MC, van Putten JP, Roos N, Aas FE, Egge-Jacobsen WM, Maier B, Koomey M. Winther-Larsen HC, et al. J Bacteriol. 2007 Sep;189(18):6676-85. doi: 10.1128/JB.00407-07. Epub 2007 Jun 15. J Bacteriol. 2007. PMID: 17573479 Free PMC article. - How Bacteria Use Type IV Pili Machinery on Surfaces.
Maier B, Wong GCL. Maier B, et al. Trends Microbiol. 2015 Dec;23(12):775-788. doi: 10.1016/j.tim.2015.09.002. Epub 2015 Oct 22. Trends Microbiol. 2015. PMID: 26497940 Review. - The type-4 pilus is the major virulence-associated adhesin of Pseudomonas aeruginosa--a review.
Hahn HP. Hahn HP. Gene. 1997 Jun 11;192(1):99-108. doi: 10.1016/s0378-1119(97)00116-9. Gene. 1997. PMID: 9224879 Review.
Cited by
- A type IV pilus mediates DNA binding during natural transformation in Streptococcus pneumoniae.
Laurenceau R, Péhau-Arnaudet G, Baconnais S, Gault J, Malosse C, Dujeancourt A, Campo N, Chamot-Rooke J, Le Cam E, Claverys JP, Fronzes R. Laurenceau R, et al. PLoS Pathog. 2013;9(6):e1003473. doi: 10.1371/journal.ppat.1003473. Epub 2013 Jun 27. PLoS Pathog. 2013. PMID: 23825953 Free PMC article. - Interactions in bacterial biofilm development: a structural perspective.
Garnett JA, Matthews S. Garnett JA, et al. Curr Protein Pept Sci. 2012 Dec;13(8):739-55. doi: 10.2174/138920312804871166. Curr Protein Pept Sci. 2012. PMID: 23305361 Free PMC article. Review. - The selective advantage of microbial fratricide.
Gilmore MS, Haas W. Gilmore MS, et al. Proc Natl Acad Sci U S A. 2005 Jun 14;102(24):8401-2. doi: 10.1073/pnas.0503828102. Epub 2005 Jun 6. Proc Natl Acad Sci U S A. 2005. PMID: 15939890 Free PMC article. No abstract available. - The molecular basis of FimT-mediated DNA uptake during bacterial natural transformation.
Braus SAG, Short FL, Holz S, Stedman MJM, Gossert AD, Hospenthal MK. Braus SAG, et al. Nat Commun. 2022 Mar 4;13(1):1065. doi: 10.1038/s41467-022-28690-1. Nat Commun. 2022. PMID: 35246533 Free PMC article. - The Role of Extracellular DNA in Microbial Attachment to Oxidized Silicon Surfaces in the Presence of Ca2+ and Na.
Morales-García AL, Walton R, Blakeman JT, Banwart SA, Harding JH, Geoghegan M, Freeman CL, Rolfe SA. Morales-García AL, et al. Langmuir. 2021 Aug 4;37(32):9838-50. doi: 10.1021/acs.langmuir.1c01410. Online ahead of print. Langmuir. 2021. PMID: 34347486 Free PMC article.
References
- Aas, F. E., M. Wolfgang, S. Frye, S. Dunham, C. Løvold, and M. Koomey. 2002. Competence for natural transformation in Neisseria gonorrhoeae: components of DNA binding and uptake linked to type IV pilus expression. Mol. Microbiol. 46:749-760. - PubMed
- Audette, G. F., R. T. Irvin, and B. Hazes. 2004. Crystallographic analysis of the Pseudomonas aeruginosa strain K122-4 monomeric pilin reveals a conserved receptor binding architecture. Biochemistry 43:11427-11435. - PubMed
- Audette, G. F., E. J. van Schaik, B. Hazes, and R. T. Irvin. Posting date, 9 September 2004. DNA-binding protein nanotubes: learning from nature's nanotech examples. Nano Lett. 4:1897-1902.[Online.]
- Bradley, D. E. 1974. The adsorption of Pseudomonas aeruginosa pilus-dependent bacteriophage to a host mutant with non-retractile pili. Virology 58:149-163. - PubMed
- Bradley, D. E., and T. L. Pitt. 1974. Pilus dependence of four Pseudomonas aeruginosa bacteriophages with non-contractile tails. J. Virol. 24:1-15. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources