Genetic diversity and population structure of teosinte - PubMed (original) (raw)
Genetic diversity and population structure of teosinte
Kenji Fukunaga et al. Genetics. 2005 Apr.
Abstract
The teosintes, the closest wild relatives of maize, are important resources for the study of maize genetics and evolution and for plant breeding. We genotyped 237 individual teosinte plants for 93 microsatellites. Phylogenetic relationships among species and subspecific taxa were largely consistent with prior analyses for other types of molecular markers. Plants of all species formed monophyletic clades, although relationships among species were not fully resolved. Phylogenetic analysis indicated that the Mexican annual teosintes divide into two clusters that largely correspond to the previously defined subspecies, Z. mays ssp. parviglumis and ssp. mexicana, although there are a few samples that represent either evolutionary intermediates or hybrids between these two subspecies. The Mexican annual teosintes show genetic substructuring along geographic lines. Hybridization or introgression between some teosintes and maize occurs at a low level and appears most common with Z. mays ssp. mexicana. Phylogeographic and phylogenetic analyses of the Mexican annual teosintes indicated that ssp. parviglumis diversified in the eastern part of its distribution and spread from east to west and that ssp. mexicana diversified in the Central Plateau of Mexico and spread along multiple paths to the north and east. We defined core sets of collections of Z. mays ssp. mexicana and ssp. parviglumis that attempt to capture the maximum number of microsatellite alleles for given sample sizes.
Figures
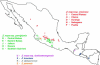
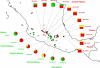
Figure 1.—
Geographical distribution of the teosinte populations used in this study. Since many accessions come from geographically very close locations, their symbols overlap on the map.
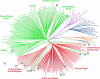
Figure 2.—
Unrooted phylogeny of individual teosinte plants using the Fitch-Margoliash method and the log-transformed proportion of shared-allele distance among 93 microsatellite loci. The tree contains 237 individuals. A large H indicates plants identified as being of putative hybrid origin by population structure analysis. Z. mays ssp. parviglumis: (B) Central Balsas, (E) eastern Balsas, (J) Jalisco, (O) Oaxaca, (S) South Guerrero. Z. mays ssp. mexicana: (C) Central Plateau, (H) Chalco, (D) Durango, (N) Nobogame, (P) Puebla. (U) Z. mays ssp. huehuetenangensis, (Xg) Z. luxurians (Guatemala), (Xn) Z. luxurians (Nicaragua), (R) Z. diploperennis, (Z) Z. perennis.
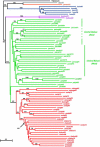
Figure 3.—
Rooted phylogeny for 76 groups (OTUs) of individual plants using the Fitch-Margoliash method and the log-transformed proportion of shared-allele distance among 93 microsatellite loci. The numbers on the branches indicate the number of times a clade appeared in 1000 bootstrap samples and are shown for all clades with >50% bootstrap support. To locate the root for Zea, separate analyses including the outgroup, Tripsacum, but only 61 SSRs, were performed. The tree as drawn shows the placement of the root when null alleles in Tripsacum are coded as distinct (not identical by descent) from null alleles in Zea (see text). Bootstrap values from this analysis are underlined.
Figure 4.—
Dispersal models for 63 OTUs of Z. mays ssp. mexicana and ssp. parviglumis. Gray circles indicate nodes of the tree, which are labeled A–F. Model 1 shows the highest correlation with genetic distance.
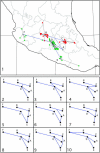
Figure 5.—
Results of the population structure analysis for ssp. mexicana (squares) and ssp. parviglumis (circles). The symbols for plants of apparent mixed ancestry based on the arbitrary criterion of possessing <80% membership in their own subspecies are enlarged. For the enlarged symbols, the symbol is color coded proportionally to its degree of ancestry from ssp. mexicana (red), ssp. parviglumis (green), and ssp. mays (yellow).
Similar articles
- Ecogeography of teosinte.
Sánchez González JJ, Ruiz Corral JA, García GM, Ojeda GR, Larios LC, Holland JB, Medrano RM, García Romero GE. Sánchez González JJ, et al. PLoS One. 2018 Feb 16;13(2):e0192676. doi: 10.1371/journal.pone.0192676. eCollection 2018. PLoS One. 2018. PMID: 29451888 Free PMC article. - Spontaneous hybridization between maize and teosinte.
Ellstrand NC, Garner LC, Hegde S, Guadagnuolo R, Blancas L. Ellstrand NC, et al. J Hered. 2007 Mar-Apr;98(2):183-7. doi: 10.1093/jhered/esm002. Epub 2007 Mar 30. J Hered. 2007. PMID: 17400586 - High segregation distortion in maize B73 x teosinte crosses.
Wang G, He QQ, Xu ZK, Song RT. Wang G, et al. Genet Mol Res. 2012 Mar 19;11(1):693-706. doi: 10.4238/2012.March.19.3. Genet Mol Res. 2012. PMID: 22535405 - The genetics of maize evolution.
Doebley J. Doebley J. Annu Rev Genet. 2004;38:37-59. doi: 10.1146/annurev.genet.38.072902.092425. Annu Rev Genet. 2004. PMID: 15568971 Review. - Genomic screening for artificial selection during domestication and improvement in maize.
Yamasaki M, Wright SI, McMullen MD. Yamasaki M, et al. Ann Bot. 2007 Nov;100(5):967-73. doi: 10.1093/aob/mcm173. Epub 2007 Aug 18. Ann Bot. 2007. PMID: 17704539 Free PMC article. Review.
Cited by
- Maize x Teosinte Hybrid Cobs Do Not Prevent Crop Gene Introgression.
Chavez NB, Flores JJ, Martin J, Ellstrand NC, Guadagnuolo R, Heredia S, Welles SR. Chavez NB, et al. Econ Bot. 2012 Jun;66(2):132-137. doi: 10.1007/s12231-012-9195-2. Epub 2012 Apr 26. Econ Bot. 2012. PMID: 22707759 Free PMC article. - Disentangling the effects of geographic and ecological isolation on genetic differentiation.
Bradburd GS, Ralph PL, Coop GM. Bradburd GS, et al. Evolution. 2013 Nov;67(11):3258-73. doi: 10.1111/evo.12193. Epub 2013 Jul 24. Evolution. 2013. PMID: 24102455 Free PMC article. - Adaptive introgression from maize has facilitated the establishment of teosinte as a noxious weed in Europe.
Le Corre V, Siol M, Vigouroux Y, Tenaillon MI, Délye C. Le Corre V, et al. Proc Natl Acad Sci U S A. 2020 Oct 13;117(41):25618-25627. doi: 10.1073/pnas.2006633117. Epub 2020 Sep 28. Proc Natl Acad Sci U S A. 2020. PMID: 32989136 Free PMC article. - Genomics of Long- and Short-Term Adaptation in Maize and Teosintes.
Lorant A, Ross-Ibarra J, Tenaillon M. Lorant A, et al. Methods Mol Biol. 2020;2090:289-311. doi: 10.1007/978-1-0716-0199-0_12. Methods Mol Biol. 2020. PMID: 31975172 Review. - Population Genomics of the Maize Pathogen Ustilago maydis: Demographic History and Role of Virulence Clusters in Adaptation.
Schweizer G, Haider MB, Barroso GV, Rössel N, Münch K, Kahmann R, Dutheil JY. Schweizer G, et al. Genome Biol Evol. 2021 May 7;13(5):evab073. doi: 10.1093/gbe/evab073. Genome Biol Evol. 2021. PMID: 33837781 Free PMC article.
References
- Benz, B. F., L. R. Sánchez-Velásquez and F. J. Santana-Michel, 1990. Ecology and ethnobotany of Zea diploperennis: preliminary investigations. Maydica 35: 85–98.
- Brown, A. H. D., 1989. Core collections: a practical approach to genetic resource management. Genome 31: 818–824.
- Buckler, E. S., IV, 1999 Phylogeographer: A tool for developing and testing phylogeographic hypotheses. http://www.maizegenetics.net/bioinformatics/phyloindex.htm.
- Buckler, E. S., IV, and T. P. Holtsford, 1996. Zea systematics: ribosomal ITS evidence. Mol. Biol. Evol. 13: 612–622. - PubMed
- Dietz, E. J., 1983. Permutation tests for association between two distance matrices. Syst. Zool. 32: 21–26.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources