ProbCons: Probabilistic consistency-based multiple sequence alignment - PubMed (original) (raw)
Comparative Study
ProbCons: Probabilistic consistency-based multiple sequence alignment
Chuong B Do et al. Genome Res. 2005 Feb.
Abstract
To study gene evolution across a wide range of organisms, biologists need accurate tools for multiple sequence alignment of protein families. Obtaining accurate alignments, however, is a difficult computational problem because of not only the high computational cost but also the lack of proper objective functions for measuring alignment quality. In this paper, we introduce probabilistic consistency, a novel scoring function for multiple sequence comparisons. We present ProbCons, a practical tool for progressive protein multiple sequence alignment based on probabilistic consistency, and evaluate its performance on several standard alignment benchmark data sets. On the BAliBASE, SABmark, and PREFAB benchmark alignment databases, ProbCons achieves statistically significant improvement over other leading methods while maintaining practical speed. ProbCons is publicly available as a Web resource.
Figures
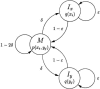
Figure 1.
Basic pair-HMM for sequence alignment between two sequences, x and y. State M emits two letters, one from each sequence, and corresponds to the two letters being aligned together. State Ix emits a letter in sequence x that is aligned to a gap, and similarly state Iy emits a letter in sequence y that is aligned to a gap. Finding the most likely alignment according to this model by using the Viterbi algorithm corresponds to applying Needleman-Wunsch with appropriate parameters. The logarithm of the emission probability function p(.,.) at M corresponds to a substitution scoring matrix, while affine gap penalty parameters can be derived from the transition probabilities δ and ε (Durbin et al. 1998).
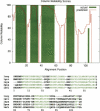
Figure 2.
Column reliability plot for 1csy_ref1 from BAliBASE, Reference 1. The red line and solid regions indicate the predicted and actual proportion of correct pairwise matches at each alignment position, respectively. All column reliability values have been multiplied by 100. Below, the actual ProbCons alignment is shown with core block residues highlighted in green. Note that only pairwise matches in core block regions of the BAliBASE alignment are considered correct when computing the “actual” proportion of correct pairwise matches; however, some residues outside of the core block regions may also be alignable. Thus, regions in which predicted homology exceeds actual homology do not necessarily indicate overprediction of homology by the aligner.
Similar articles
- OXBench: a benchmark for evaluation of protein multiple sequence alignment accuracy.
Raghava GP, Searle SM, Audley PC, Barber JD, Barton GJ. Raghava GP, et al. BMC Bioinformatics. 2003 Oct 10;4:47. doi: 10.1186/1471-2105-4-47. BMC Bioinformatics. 2003. PMID: 14552658 Free PMC article. - MSAProbs: multiple sequence alignment based on pair hidden Markov models and partition function posterior probabilities.
Liu Y, Schmidt B, Maskell DL. Liu Y, et al. Bioinformatics. 2010 Aug 15;26(16):1958-64. doi: 10.1093/bioinformatics/btq338. Epub 2010 Jun 23. Bioinformatics. 2010. PMID: 20576627 - A knowledge-based multiple-sequence alignment algorithm.
Nguyen KD, Pan Y. Nguyen KD, et al. IEEE/ACM Trans Comput Biol Bioinform. 2013 Jul-Aug;10(4):884-96. doi: 10.1109/TCBB.2013.102. IEEE/ACM Trans Comput Biol Bioinform. 2013. PMID: 24334383 - Multiple sequence alignment.
Edgar RC, Batzoglou S. Edgar RC, et al. Curr Opin Struct Biol. 2006 Jun;16(3):368-73. doi: 10.1016/j.sbi.2006.04.004. Epub 2006 May 5. Curr Opin Struct Biol. 2006. PMID: 16679011 Review. - Upcoming challenges for multiple sequence alignment methods in the high-throughput era.
Kemena C, Notredame C. Kemena C, et al. Bioinformatics. 2009 Oct 1;25(19):2455-65. doi: 10.1093/bioinformatics/btp452. Epub 2009 Jul 30. Bioinformatics. 2009. PMID: 19648142 Free PMC article. Review.
Cited by
- Three duplication events and variable molecular evolution characteristics involved in multiple GGPS genes of six Solanaceae species.
Li F, Wei CY, Qiao C, Chen Z, Wang P, Wei P, Wang R, Jin L, Yang J, Lin F, Luo Z. Li F, et al. J Genet. 2016 Jun;95(2):453-7. doi: 10.1007/s12041-016-0634-1. J Genet. 2016. PMID: 27350691 No abstract available. - Dynamically evolving novel overlapping gene as a factor in the SARS-CoV-2 pandemic.
Nelson CW, Ardern Z, Goldberg TL, Meng C, Kuo CH, Ludwig C, Kolokotronis SO, Wei X. Nelson CW, et al. Elife. 2020 Oct 1;9:e59633. doi: 10.7554/eLife.59633. Elife. 2020. PMID: 33001029 Free PMC article. - The Escherichia coli RlmN methyltransferase is a dual-specificity enzyme that modifies both rRNA and tRNA and controls translational accuracy.
Benítez-Páez A, Villarroya M, Armengod ME. Benítez-Páez A, et al. RNA. 2012 Oct;18(10):1783-95. doi: 10.1261/rna.033266.112. Epub 2012 Aug 13. RNA. 2012. PMID: 22891362 Free PMC article. - GUIDANCE2: accurate detection of unreliable alignment regions accounting for the uncertainty of multiple parameters.
Sela I, Ashkenazy H, Katoh K, Pupko T. Sela I, et al. Nucleic Acids Res. 2015 Jul 1;43(W1):W7-14. doi: 10.1093/nar/gkv318. Epub 2015 Apr 16. Nucleic Acids Res. 2015. PMID: 25883146 Free PMC article. - Structural and molecular basis of interaction of HCV non-structural protein 5A with human casein kinase 1α and PKR.
Sudha G, Yamunadevi S, Tyagi N, Das S, Srinivasan N. Sudha G, et al. BMC Struct Biol. 2012 Nov 13;12:28. doi: 10.1186/1472-6807-12-28. BMC Struct Biol. 2012. PMID: 23148689 Free PMC article.
References
- Altschul, S.F., Carroll, R.J., and Lipman, D.J. 1989. Weights for data related by a tree. J. Mol. Biol. 207: 647-653. - PubMed
- Attwood, T.K. 2002. The PRINTS database: A resource for identification of protein families. Brief. Bioinform. 3: 252-263. - PubMed
WEB SITE REFERENCES
- http://probcons.stanford.edu; ProbCons alignment tool.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources