Accelerated evolution associated with genome reduction in a free-living prokaryote - PubMed (original) (raw)
Accelerated evolution associated with genome reduction in a free-living prokaryote
Alexis Dufresne et al. Genome Biol. 2005.
Abstract
Background: Three complete genomes of Prochlorococcus species, the smallest and most abundant photosynthetic organism in the ocean, have recently been published. Comparative genome analyses reveal that genome shrinkage has occurred within this genus, associated with a sharp reduction in G+C content. As all examples of genome reduction characterized so far have been restricted to endosymbionts or pathogens, with a host-dependent lifestyle, the observed genome reduction in Prochlorococcus is the first documented example of such a process in a free-living organism.
Results: Our results clearly indicate that genome reduction has been accompanied by an increased rate of protein evolution in P. marinus SS120 that is even more pronounced in P. marinus MED4. This acceleration has affected every functional category of protein-coding genes. In contrast, the 16S rRNA gene seems to have evolved clock-like in this genus. We observed that MED4 and SS120 have lost several DNA-repair genes, the absence of which could be related to the mutational bias and the acceleration of amino-acid substitution.
Conclusions: We have examined the evolutionary mechanisms involved in this process, which are different from those known from host-dependent organisms. Indeed, most substitutions that have occurred in Prochlorococcus have to be selectively neutral, as the large size of populations imposes low genetic drift and strong purifying selection. We assume that the major driving force behind genome reduction within the Prochlorococcus radiation has been a selective process favoring the adaptation of this organism to its environment. A scenario is proposed for genome evolution in this genus.
Figures
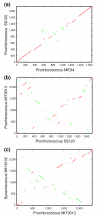
Figure 1
Alignments of complete genome sequences of marine picocyanobacteria. Genome sequences are translated in their six reading frames. (a) Comparison of the MED4 and SS120 genomes; (b) comparison of the SS120 and MIT9313 genomes; (c) comparison of the MIT9313 and WH8102 genomes. Colinear segments are shown in red and inversions in green. Translocated segments are above or below the diagonal.
Figure 2
Phylogenetic tree of 16S rRNA genes from the four marine picocyanobacteria. Neighbor-joining tree with Kimura 2-parameter correction. The bootstrap value (1,000 replications) is shown in boldface. Lengths of the branches dA, dB, dC and dX (see text) are given below the branches. N1, node 1, branchpoint between MED4 and SS120; N2, node 2, branchpoint between MIT9313 and Node 1.
Figure 3
Influence of mutational bias in codon usage and amino-acid usage. (a) Percentage use of AT-rich codons in the four marine picocyanobacteria. Amino acids are ranked according to AT content of their respective codons. Methionine and tryptophan, which are both encoded by only one codon, have been discarded from the analysis. (b) Percentage use of amino acids in marine picocyanobacteria.
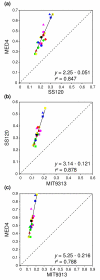
Figure 4
Amino-acid substitution rate per functional category. Branch lengths computed for each functional category between (a) MED4 and SS120, (b) SS120 and MIT9313 and (c) MED4 and MIT9313. In the three comparisons, branch-length values are aligned along a line with a slope much greater than 1, indicating that acceleration of the substitution rates occurs in every functional category. Axes represent the number of amino-acid substitutions per site. Red circle, amino-acid transport and metabolism; green circle, carbohydrate transport and metabolism; yellow circle, cell-cycle control; blue triangle, cell wall/membrane biogenesis; pink circle, coenzyme metabolism; red square, defense mechanisms; green square, energy production and conversion; yellow square, function unknown; blue square, general function prediction only; pink square, inorganic ion transport and metabolism; red triangle, intracellular trafficking; green triangle, lipid transport and metabolism; yellow triangle, nucleotide transport and metabolism; blue circle, posttranslational modification, protein turnover; pink triangle, replication, recombination and repair; red diamond, secondary metabolite biosynthesis, transport and catabolism; green diamond, signal transduction mechanisms; yellow diamond, transcription; blue diamond, translation; black circle, miscellaneous.
References
- Lawrence JG, Roth JR. Genomic flux: genome evolution by gene loss and acquisition. In: Charlebois RL, editor. Organization of the Prokaryotic Genome. Washington, DC: American Society for Microbiology; 1999. pp. 263–289.
- Fraser CM, Gocayne JD, White O, Adams MD, Clayton RA, Fleischmann RD, Bult CJ, Kerlavage AR, Sutton G, Kelley JM, et al. The minimal gene complement of Mycoplasma genitalium. Science. 1995;270:397–403. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials