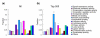Identification of ciliated sensory neuron-expressed genes in Caenorhabditis elegans using targeted pull-down of poly(A) tails - PubMed (original) (raw)
Identification of ciliated sensory neuron-expressed genes in Caenorhabditis elegans using targeted pull-down of poly(A) tails
Hirofumi Kunitomo et al. Genome Biol. 2005.
Abstract
It is not always easy to apply microarray technology to small numbers of cells because of the difficulty in selectively isolating mRNA from such cells. We report here the preparation of mRNA from ciliated sensory neurons of Caenorhabditis elegans using the mRNA-tagging method, in which poly(A) RNA was co-immunoprecipitated with an epitope-tagged poly(A)-binding protein specifically expressed in sensory neurons. Subsequent cDNA microarray analyses led to the identification of a panel of sensory neuron-expressed genes.
Figures
Figure 1
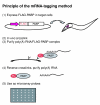
Principle of the mRNA-tagging method. Step 1, FLAG-tagged poly(A)-binding protein (PABP) is expressed from a transgene using a cell-specific promoter. Step 2, PABP and poly(A)+ RNA are crosslinked in situ by formaldehyde. Step 3, poly(A)-RNA/FLAG-PABP complexes are purified by anti-FLAG affinity purification. Step 4, RNA-PABP crosslinks are reversed and RNA is isolated. Step 5, purified RNA is used for microarray analysis.
Figure 2
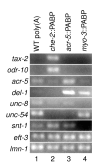
Quantification of tissue-specific transcripts in RNA prepared by mRNA tagging. The transcript indicated on the left of each row was amplified by RT-PCR using gene-specific primers. Poly(A)+RNA from wild-type (WT) animals was used as a template in lane 1. RNA prepared by mRNA tagging from che-2::PABP (JN501), acr-5::PABP (JN502) and myo-3::PABP (JN503) was used in lanes 2, 3 and 4, respectively.
Figure 3
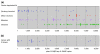
Rank orders of che-2::PABP/acr-5::PABP values for specific genes in the microarray analyses. (a) Distribution of genes with known expression patterns. Genes known to be specifically expressed in sensory neurons, motor neurons, muscles or the intestine, respectively, were collected from WormBase (see Materials and methods) and the rank orders of their che-2::PABP/acr-5::PABP signal ratios were plotted. Vertical bars indicate the medians. Genes expressed in sensory neurons are specifically enriched in the che-2::PABP RNA preparations, while motor neuron- and intestine-expressed genes are enriched in the acr-5::PABP RNA preparations. Note that although only five genes were found as motor neuron-expressed genes, nine data points were plotted in (a), because multiple cDNA clones were present on the microarray for three of the genes (see Additional data file 2). (b) Distribution of genes with X-boxes in their promoter regions. Genes that carry one or more X-boxes in their promoter regions were collected from the genome database (see Materials and methods) and their rank orders of che-2::PABP/acr-5::PABP signal ratios were plotted. These genes, which are expected to be expressed in ciliated sensory neurons under the control of the DAF-19 transcription factor, are also enriched in the che-2::PABP RNA preparations.
Figure 4
Expression patterns of newly identified sensory neuron-expressed genes. The genes indicated were each fused to GFP in-frame, and the reporters introduced into wild-type animals. Overlaid images of the Nomarski and GFP fluorescence images of transgenic worms between larval stages 1 and 3 are shown. Gene expression is indicated by the green fluorescence. Scale bar, 50 μm. See Table 1 for the identity of the expressing cells.
Figure 5
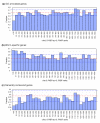
Sensory neuron-specific genes are less likely to be classified into Gene Ontology categories and more likely to be worm-specific. (a) All genes on the microarray were ordered by descending che-2::PABP/acr-5::PABP value and the fraction of GO-annotated genes in each bin is indicated for a bin width of 50 rank orders. Only the top 1,500 genes are shown in (a)-(c). (b) The fraction of genes with homologs in C. briggsae, and not in humans, mice, flies, fission yeast or budding yeast (cutoff BLASTP score E = 1 × 10-20) in each bin is indicated as in (a). (c) The fraction of genes with homologs in both animals and yeasts, namely in humans, mice or flies and in fission yeast or budding yeast (cutoff BLASTP score E = 1 × 10-20) in each bin is indicated as in (a). In all panels, the red dotted line indicates the average of all the genes, and the blue dotted lines indicate the 95% confidence limits assuming a random binominal distribution.
Figure 6
Categories of genes enriched in the sensory neuron fraction. Genes were categorized according to the GO molecular function categories. (a) Categorization of all the genes on the microarray; (b) categorization of genes within the top 500 che-2::PABP/acr-5::PABP ranks. In both panels, the fraction of genes in each category in respect of all annotated genes is shown.*P < 0.05; **P < 0.01 (binominal distribution).
Similar articles
- Isolation of mRNA from specific tissues of Drosophila by mRNA tagging.
Yang Z, Edenberg HJ, Davis RL. Yang Z, et al. Nucleic Acids Res. 2005 Oct 4;33(17):e148. doi: 10.1093/nar/gni149. Nucleic Acids Res. 2005. PMID: 16204451 Free PMC article. - Identification of genes expressed in C. elegans touch receptor neurons.
Zhang Y, Ma C, Delohery T, Nasipak B, Foat BC, Bounoutas A, Bussemaker HJ, Kim SK, Chalfie M. Zhang Y, et al. Nature. 2002 Jul 18;418(6895):331-5. doi: 10.1038/nature00891. Nature. 2002. PMID: 12124626 - Cell-specific microarray profiling experiments reveal a comprehensive picture of gene expression in the C. elegans nervous system.
Von Stetina SE, Watson JD, Fox RM, Olszewski KL, Spencer WC, Roy PJ, Miller DM 3rd. Von Stetina SE, et al. Genome Biol. 2007;8(7):R135. doi: 10.1186/gb-2007-8-7-r135. Genome Biol. 2007. PMID: 17612406 Free PMC article. - Profiling C. elegans gene expression with DNA microarrays.
Portman DS. Portman DS. WormBook. 2006 Jan 20:1-11. doi: 10.1895/wormbook.1.104.1. WormBook. 2006. PMID: 18050445 Free PMC article. Review. No abstract available. - Reporter gene fusions.
Boulin T, Etchberger JF, Hobert O. Boulin T, et al. WormBook. 2006 Apr 5:1-23. doi: 10.1895/wormbook.1.106.1. WormBook. 2006. PMID: 18050449 Free PMC article. Review. No abstract available.
Cited by
- The relationship between intraflagellar transport and upstream protein trafficking pathways and macrocyclic lactone resistance in Caenorhabditis elegans.
Brinzer RA, Winter AD, Page AP. Brinzer RA, et al. G3 (Bethesda). 2024 Mar 6;14(3):jkae009. doi: 10.1093/g3journal/jkae009. G3 (Bethesda). 2024. PMID: 38227795 Free PMC article. - Tissue-Specific Transcription Footprinting Using RNA PoI DamID (RAPID) in Caenorhabditis elegans.
Gómez-Saldivar G, Osuna-Luque J, Semple JI, Glauser DA, Jarriault S, Meister P. Gómez-Saldivar G, et al. Genetics. 2020 Dec;216(4):931-945. doi: 10.1534/genetics.120.303774. Epub 2020 Oct 9. Genetics. 2020. PMID: 33037050 Free PMC article. - The sensory cilia of Caenorhabditis elegans.
Inglis PN, Ou G, Leroux MR, Scholey JM. Inglis PN, et al. WormBook. 2007 Mar 8:1-22. doi: 10.1895/wormbook.1.126.2. WormBook. 2007. PMID: 18050505 Free PMC article. Review. - Identification of ciliary and ciliopathy genes in Caenorhabditis elegans through comparative genomics.
Chen N, Mah A, Blacque OE, Chu J, Phgora K, Bakhoum MW, Newbury CR, Khattra J, Chan S, Go A, Efimenko E, Johnsen R, Phirke P, Swoboda P, Marra M, Moerman DG, Leroux MR, Baillie DL, Stein LD. Chen N, et al. Genome Biol. 2006;7(12):R126. doi: 10.1186/gb-2006-7-12-r126. Genome Biol. 2006. PMID: 17187676 Free PMC article. - Chromosomal clustering and GATA transcriptional regulation of intestine-expressed genes in C. elegans.
Pauli F, Liu Y, Kim YA, Chen PJ, Kim SK. Pauli F, et al. Development. 2006 Jan;133(2):287-95. doi: 10.1242/dev.02185. Epub 2005 Dec 14. Development. 2006. PMID: 16354718 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources