RNA interference rescue by bacterial artificial chromosome transgenesis in mammalian tissue culture cells - PubMed (original) (raw)
RNA interference rescue by bacterial artificial chromosome transgenesis in mammalian tissue culture cells
Ralf Kittler et al. Proc Natl Acad Sci U S A. 2005.
Abstract
RNA interference (RNAi) is a widely used method for analysis of gene function in tissue culture cells. However, to date there has been no reliable method for testing the specificity of any particular RNAi experiment. The ideal experiment is to rescue the phenotype by expression of the target gene in a form refractory to RNAi. The transgene should be expressed at physiological levels and with its different splice variants. Here, we demonstrate that expression of murine bacterial artificial chromosomes in human cells provides a reliable method to create RNAi-resistant transgenes. This strategy should be applicable to all eukaryotes and should therefore be a standard technology for confirming the specificity of RNAi. We show that this technique can be extended to allow the creation of tagged transgenes, expressed at physiological levels, for the further study of gene function.
Figures
Fig. 1.
Experimental strategy of RNAi rescue by BAC transgenesis. RFP, red fluorescent protein; Kan/neor, kanamycin/neomycin resistance gene; Cmr, chloramphenicol resistance gene; ori, origin of replication; BB, BAC backbone; CSI-BAC, cross-species RNAi rescue-BAC.
Fig. 2.
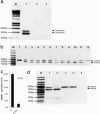
Expression and RNAi resistance of mouse transgenes. (a) Detection of expression and alternative splicing of mDNAJA3 by RT-PCR. Lane 1, the two bands (495-bp and 377-bp) amplified from cDNA of HeLa cells transfected and selected for a BAC carrying mDNAJA3 representing the two splice isoforms; lane 2, negative control (cDNA from WT HeLa cells); lane 3, non-template control. M, marker. (b) Expression levels of hSNW1 and mSNW1 in transgenic cells. The comparison of the band intensities indicates relative expression levels of hSNW1 and mSNW1. Lane 1, undigested 161-bp fragment from the same clone depicted in lane 2; lanes 2–11, Sfa_NI digestion products of a 161-bp fragment amplified from cDNA of 10 clones from HeLa cells transfected and selected for a BAC carrying mSNW1 (the upper band represents the uncut human-specific fragment, the lower band the mouse-specific fragment); lane 12, digested fragment generated from cDNA of WT HeLa cells. M, Marker. (c) Knockdown of hSNW1 in transgenic HeLa cells. hSNW1 expression after transfection of esiRNAs targeting hSNW1 and firefly luciferase. hSNW1 mRNA expression was quantified 2 days after transfection. Expression levels were normalized against h_SNW1 expression of cells transfected with esiRNA targeting firefly luciferase. (d) Expression levels of hSNW1 and mSNW1 in transgenic HeLa cells. Lanes 1 and 2, _Sfa_NI digestion products of the 161-bp fragment amplified from cDNA of transgenic HeLa cells transfected with esiRNA targeting firefly luciferase (lane 1) and hSNW1 (lane 2) (note the change of relative band intensities indicating the specific knockdown of hSNW1); lanes 3 and 4, undigested 161-bp fragment amplified from cDNA of transgenic HeLa cells transfected with esiRNA targeting firefly luciferase (lane 3) and hSNW1 (lane 2); lane 5, non-template control. M, marker.
Fig. 3.
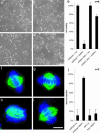
Rescue experiment. (a–d) Phase contrast microscopic images of WT HeLa cells (a and c) and BAC transgenic HeLa cells expressing mSNW1 (b and d)96h after transfection with esiRNA targeting hSNW1 (c and d) and firefly luciferase as negative control (a and b). (e) Effect on cell viability of esiRNA targeting hSNW1. Cells were assayed 96 h after transfection. Shown is the reduction of cell viability as determined with the WST-1 assay normalized against the negative control luc (esiRNA directed against firefly luciferase). (f–i) WT HeLa cells (f and h) and BAC transgenic HeLa cells expressing mSNW1 (g and i) were transfected with esiRNA targeting hSNW1 (h and i) and firefly luciferase as negative control (f and g) and imaged by 3D deconvolution microscopy 48 h after transfection for tubulin (green) and DNA (blue). [Scale bars: 100 μm(a–d) and 5 μm(f–i).] (j) Frequency of aberrant spindles. We counted the frequency of aberrant spindles 48 h after transfection for 50 mitotic cells per coverslip. Error bars represent the standard variation. mSNW1-BAC, HeLa cells that have stably integrated the BAC carrying mSNW1; luc, esiRNA targeting firefly luciferase; hSNW1, esiRNA targeting hSNW1.
Fig. 4.
BAC tagging for functional studies in mammalian cell lines. (a) Diagram illustrating procedures. TAG, fusion sequence to investigate gene function (e.g., GFP); IRES, internal ribosome entry site; neo, kanamycin/neomycin resistance gene; r, resistance gene of the BAC; BB, BAC backbone. (b) HeLa cell line containing the mouse SPD2 BAC tagged with GFP stained for DNA (blue), microtubules (red), and GFP (green). Note the presence of mSPD2-GFP on both poles of the mitotic spindle. (c) WT HeLa cells or HeLa cells stably expressing the mSPD2 BAC tagged with GFP submitted to hSPD2 RNAi. Error bars represent the standard variation. Note that the percentage of mitotic cells present in HeLa mSPD2 BAC-containing cells is reduced compared with that observed in WT cells.
Similar articles
- The best control for the specificity of RNAi.
Sarov M, Stewart AF. Sarov M, et al. Trends Biotechnol. 2005 Sep;23(9):446-8. doi: 10.1016/j.tibtech.2005.06.007. Trends Biotechnol. 2005. PMID: 15979179 Review. - Reconstitution of CKMT1 expression fails to rescue cells from mitochondrial membrane potential dissipation: implications for controlling RNAi experiments.
Datler C, Grimm S. Datler C, et al. Biochim Biophys Acta. 2013 Dec;1833(12):2844-2855. doi: 10.1016/j.bbamcr.2013.07.006. Epub 2013 Jul 21. Biochim Biophys Acta. 2013. PMID: 23880370 - A tightly regulated and reversibly inducible siRNA expression system for conditional RNAi-mediated gene silencing in mammalian cells.
Wu RH, Cheng TL, Lo SR, Hsu HC, Hung CF, Teng CF, Wu MP, Tsai WH, Chang WT. Wu RH, et al. J Gene Med. 2007 Jul;9(7):620-34. doi: 10.1002/jgm.1048. J Gene Med. 2007. PMID: 17486668 - A recombineering pipeline for functional genomics applied to Caenorhabditis elegans.
Sarov M, Schneider S, Pozniakovski A, Roguev A, Ernst S, Zhang Y, Hyman AA, Stewart AF. Sarov M, et al. Nat Methods. 2006 Oct;3(10):839-44. doi: 10.1038/nmeth933. Nat Methods. 2006. PMID: 16990816 - Nonviral vector-mediated RNA interference: its gene silencing characteristics and important factors to achieve RNAi-based gene therapy.
Takahashi Y, Nishikawa M, Takakura Y. Takahashi Y, et al. Adv Drug Deliv Rev. 2009 Jul 25;61(9):760-6. doi: 10.1016/j.addr.2009.04.006. Epub 2009 Apr 20. Adv Drug Deliv Rev. 2009. PMID: 19386274 Review.
Cited by
- Short-hairpin RNA-mediated stable silencing of Grb2 impairs cell growth and DNA synthesis.
Di Fulvio M, Henkels KM, Gomez-Cambronero J. Di Fulvio M, et al. Biochem Biophys Res Commun. 2007 Jun 8;357(3):737-42. doi: 10.1016/j.bbrc.2007.04.013. Epub 2007 Apr 10. Biochem Biophys Res Commun. 2007. PMID: 17445773 Free PMC article. - TPX2 regulates the localization and activity of Eg5 in the mammalian mitotic spindle.
Ma N, Titus J, Gable A, Ross JL, Wadsworth P. Ma N, et al. J Cell Biol. 2011 Oct 3;195(1):87-98. doi: 10.1083/jcb.201106149. J Cell Biol. 2011. PMID: 21969468 Free PMC article. - The Grb2/PLD2 interaction is essential for lipase activity, intracellular localization and signaling in response to EGF.
Di Fulvio M, Frondorf K, Henkels KM, Lehman N, Gomez-Cambronero J. Di Fulvio M, et al. J Mol Biol. 2007 Mar 30;367(3):814-24. doi: 10.1016/j.jmb.2007.01.021. Epub 2007 Jan 12. J Mol Biol. 2007. PMID: 17276458 Free PMC article. - Cross-species RNAi rescue platform in Drosophila melanogaster.
Kondo S, Booker M, Perrimon N. Kondo S, et al. Genetics. 2009 Nov;183(3):1165-73. doi: 10.1534/genetics.109.106567. Epub 2009 Aug 31. Genetics. 2009. PMID: 19720858 Free PMC article. - MISSION esiRNA for RNAi screening in mammalian cells.
Theis M, Buchholz F. Theis M, et al. J Vis Exp. 2010 May 12;(39):2008. doi: 10.3791/2008. J Vis Exp. 2010. PMID: 20467416 Free PMC article.
References
- Carpenter, A. E. & Sabatini, D. M. (2004) Nat. Rev. Genet. 5, 11-22. - PubMed
- Dykxhoorn, D. M., Novina, C. D. & Sharp, P. A. (2003) Nat. Rev. Mol. Cell Biol. 4, 457-467. - PubMed
- Jackson, A. L., Bartz, S. R., Schelter, J., Kobayashi, S. V., Burchard, J., Mao, M., Li, B., Cavet, G. & Linsley, P. S. (2003) Nat. Biotechnol. 21, 635-637. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources