Evaluation of a two-step approach for large-scale, prospective genotyping of Mycobacterium tuberculosis isolates in the United States - PubMed (original) (raw)
Comparative Study
Evaluation of a two-step approach for large-scale, prospective genotyping of Mycobacterium tuberculosis isolates in the United States
Lauren S Cowan et al. J Clin Microbiol. 2005 Feb.
Abstract
Genotyping of Mycobacterium tuberculosis isolates is useful in tuberculosis control for confirming suspected transmission links, identifying unsuspected transmission, and detecting or confirming possible false-positive cultures. The value is greatly increased by reducing the turnaround time from positive culture to genotyping result and by increasing the proportion of cases for which results are available. Although IS6110 fingerprinting provides the highest discrimination, amplification-based methods allow rapid, high-throughput processing and yield digital results that can be readily analyzed and thus are better suited for large-scale genotyping. M. tuberculosis isolates (n = 259) representing 99% of culture-positive cases of tuberculosis diagnosed in Wisconsin in the years 2000 to 2003 were genotyped by using spoligotyping, mycobacterial interspersed repetitive unit (MIRU) typing, and IS6110 fingerprinting. Spoligotyping clustered 64.1% of the isolates, MIRU typing clustered 46.7% of the isolates, and IS6110 fingerprinting clustered 29.7% of the isolates. The combination of spoligotyping and MIRU typing yielded 184 unique isolates and 26 clusters containing 75 isolates (29.0%). The addition of IS6110 fingerprinting reduced the number of clustered isolates to 30 (11.6%) if an exact pattern match was required or to 44 (17.0%) if the definition of a matching IS6110 fingerprint was expanded to include patterns that differed by the addition of a single band. Regardless of the genotyping method chosen, the addition of a second or third method decreased clustering. Our results indicate that using spoligotyping and MIRU typing together provides adequate discrimination in most cases. IS6110 fingerprinting can then be used as a secondary typing method to type the clustered isolates when additional discrimination is needed.
Figures
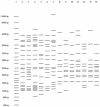
FIG. 1.
Emergence of common spoligotype-MIRU patterns. Lane 1, size markers; lanes 2 to 7, IS_6110_ fingerprints for isolates with the spoligotype 000000000003771 and MIRU pattern 223325173533 (Beijing genotype family); lanes 8 to 14, IS_6110_ fingerprints for isolates with the spoligotype 677777477413771 and MIRU pattern 254326223432 (Manila genotype family).
Similar articles
- Sensitivities and specificities of spoligotyping and mycobacterial interspersed repetitive unit-variable-number tandem repeat typing methods for studying molecular epidemiology of tuberculosis.
Scott AN, Menzies D, Tannenbaum TN, Thibert L, Kozak R, Joseph L, Schwartzman K, Behr MA. Scott AN, et al. J Clin Microbiol. 2005 Jan;43(1):89-94. doi: 10.1128/JCM.43.1.89-94.2005. J Clin Microbiol. 2005. PMID: 15634955 Free PMC article. - Secondary typing of Mycobacterium tuberculosis isolates with matching IS6110 fingerprints from different geographic regions of the United States.
Yang ZH, Bates JH, Eisenach KD, Cave MD. Yang ZH, et al. J Clin Microbiol. 2001 May;39(5):1691-5. doi: 10.1128/JCM.39.5.1691-1695.2001. J Clin Microbiol. 2001. PMID: 11325975 Free PMC article. - Assessment of an optimized mycobacterial interspersed repetitive- unit-variable-number tandem-repeat typing system combined with spoligotyping for population-based molecular epidemiology studies of tuberculosis.
Oelemann MC, Diel R, Vatin V, Haas W, Rüsch-Gerdes S, Locht C, Niemann S, Supply P. Oelemann MC, et al. J Clin Microbiol. 2007 Mar;45(3):691-7. doi: 10.1128/JCM.01393-06. Epub 2006 Dec 27. J Clin Microbiol. 2007. PMID: 17192416 Free PMC article. - Genotyping in contact investigations: a CDC perspective.
Crawford JT. Crawford JT. Int J Tuberc Lung Dis. 2003 Dec;7(12 Suppl 3):S453-7. Int J Tuberc Lung Dis. 2003. PMID: 14677837 Review. - The Evolution of Strain Typing in the Mycobacterium tuberculosis Complex.
Merker M, Kohl TA, Niemann S, Supply P. Merker M, et al. Adv Exp Med Biol. 2017;1019:43-78. doi: 10.1007/978-3-319-64371-7_3. Adv Exp Med Biol. 2017. PMID: 29116629 Review.
Cited by
- Molecular epidemiology of tuberculosis: current insights.
Mathema B, Kurepina NE, Bifani PJ, Kreiswirth BN. Mathema B, et al. Clin Microbiol Rev. 2006 Oct;19(4):658-85. doi: 10.1128/CMR.00061-05. Clin Microbiol Rev. 2006. PMID: 17041139 Free PMC article. Review. - Current methods in the molecular typing of Mycobacterium tuberculosis and other mycobacteria.
Jagielski T, van Ingen J, Rastogi N, Dziadek J, Mazur PK, Bielecki J. Jagielski T, et al. Biomed Res Int. 2014;2014:645802. doi: 10.1155/2014/645802. Epub 2014 Jan 5. Biomed Res Int. 2014. PMID: 24527454 Free PMC article. Review. - Risk factors associated with cluster size of Mycobacterium tuberculosis (Mtb) of different RFLP lineages in Brazil.
Peres RL, Vinhas SA, Ribeiro FKC, Palaci M, do Prado TN, Reis-Santos B, Zandonade E, Suffys PN, Golub JE, Riley LW, Maciel EL. Peres RL, et al. BMC Infect Dis. 2018 Feb 8;18(1):71. doi: 10.1186/s12879-018-2969-0. BMC Infect Dis. 2018. PMID: 29422032 Free PMC article. - Microevolution of the direct repeat locus of Mycobacterium tuberculosis in a strain prevalent in San Francisco.
Aga RS, Fair E, Abernethy NF, Deriemer K, Paz EA, Kawamura LM, Small PM, Kato-Maeda M. Aga RS, et al. J Clin Microbiol. 2006 Apr;44(4):1558-60. doi: 10.1128/JCM.44.4.1558-1560.2006. J Clin Microbiol. 2006. PMID: 16597893 Free PMC article. - Investigation on Mycobacterium tuberculosis diversity in China and the origin of the Beijing clade.
Wan K, Liu J, Hauck Y, Zhang Y, Liu J, Zhao X, Liu Z, Lu B, Dong H, Jiang Y, Kremer K, Vergnaud G, van Soolingen D, Pourcel C. Wan K, et al. PLoS One. 2011;6(12):e29190. doi: 10.1371/journal.pone.0029190. Epub 2011 Dec 29. PLoS One. 2011. PMID: 22220207 Free PMC article.
References
- Cronin, W. A., J. E. Golub, L. S. Magder, N. G. Baruch, M. J. Lathan, L. N. Mukasa, N. Hooper, J. H. Razeq, D. Mulcahy, W. H. Benjamin, and W. R. Bishai. 2001. Epidemiologic usefulness of spoligotyping for secondary typing of Mycobacterium tuberculosis with low copy numbers of IS6110. J. Clin. Microbiol. 39:3709-3711. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources