Hepatitis C virus RNA replication is regulated by host geranylgeranylation and fatty acids - PubMed (original) (raw)
Hepatitis C virus RNA replication is regulated by host geranylgeranylation and fatty acids
Sharookh B Kapadia et al. Proc Natl Acad Sci U S A. 2005.
Abstract
Hepatitis C virus (HCV) infection is a major cause of chronic hepatitis, liver cirrhosis, and hepatocellular carcinoma. Our laboratory has previously demonstrated that high-level HCV replication during acute infection of chimpanzees is associated with the modulation of multiple genes involved in lipid metabolism, and that drugs that regulate cholesterol and fatty acid biosynthesis regulate the replication of the subgenomic HCV replicon in Huh-7 cells. In this article, we demonstrate that Huh-7 cells harboring replicating, full-length HCV RNAs express elevated levels of ATP citrate lyase and acetyl-CoA synthetase genes, both of which are involved in cholesterol and fatty acid biosynthesis. Further, we confirm that the cholesterol-biosynthetic pathway controls HCV RNA replication by regulating the cellular levels of geranylgeranyl pyrophosphate, we demonstrate that the impact of geranylgeranylation depends on the fatty acid content of the cell, and we show that fatty acids can either stimulate or inhibit HCV replication, depending on their degree of saturation. These results illustrate a complex cellular-regulatory network that controls HCV RNA replication, presumably by modulating the trafficking and association of cellular and/or viral proteins with cellular membranes, suggesting that pharmacologic manipulation of these pathways may have a therapeutic effect in chronic HCV infection.
Figures
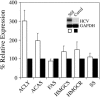
Fig. 1.
Induction of ATP citrate lyase and acetyl-CoA synthetase gene expression in Huh-7 cells that replicate the full-length SfiI replicon. RPA was performed by using probes specific to ATP citrate lyase (ACLY), acetyl-CoA synthetase (ACAS), fatty acid synthase (FAS), HMG-CoA synthase (HMGCS), HMG-CoA reductase (HMGCR), and squalene synthase (SS), and expression levels in SfiI cells (white bars) were compared with IFN-cured cells (black bars). Representative data (mean + SD) from three independent experiments are shown. The Inset represents Northern blot analysis to detect HCV RNA.
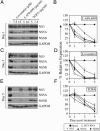
Fig. 2.
Geranylgeranylation is required for HCV RNA replication. Northern blot analyses were performed on total RNA harvested from SfiI cells treated with various small-molecule inhibitors. (A) SfiI cells were treated with either 7.5 or 15 μM lovastatin alone or in the presence of 10 μM geranylgeraniol or farnesol. (B) SfiI cells were treated with 5, 10, or 20 μg/ml L-659,699, and total RNA was harvested at 5 days posttreatment. (C) SfiI cells were treated with 20 μg/ml ZA alone or in the presence or absence of low-density lipoprotein (LDL). (D) SfiI cells were treated with 10 μM geranylgeraniol (GGOH) or farnesol (FOH), and total RNA was harvested on day 5 posttreatment. In all cases, relative levels of HCV RNA are shown as percentages normalized to GAPDH RNA levels. *, Transcript levels were undetectable.
Fig. 3.
Inhibition of HCV protein synthesis by small-molecule inhibitors of cholesterol and fatty acid biosynthesis. (A, C, and E) SfiI cells were mock-treated or treated with increasing doses of lovastatin, L-659,699, or TOFA. Total cell lysates were harvested at various times posttreatment, and SDS/PAGE analyses were performed by using antibodies specific to NS3, NS5A, NS5B, or GAPDH. (B, D, and F) Kinetic analysis of HCV protein expression of SfiI cells treated with L-659,699 (B), lovastatin (D), or TOFA (F) compared with HCV RNA levels. Representative data (mean ± SD) from at least three independent experiments are shown.
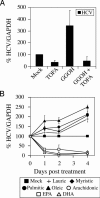
Fig. 4.
Inhibition of basal and geranylgeraniol-induced HCV RNA replication by TOFA. (A) SfiI cells were mock-treated or treated with 5 μg/ml TOFA, 10 μM geranylgeraniol (GGOH), or TOFA and GGOH. (B) SfiI cells were treated with 50 μM fatty acids complexed to fatty-acid-free BSA. Total RNA was harvested at various times posttreatment and Northern blot analysis was performed. Levels of HCV RNA are represented as a percentage normalized to GAPDH RNA levels. Representative data (mean + SD) from three independent experiments are shown.
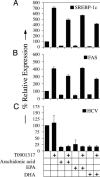
Fig. 5.
PUFA-mediated inhibition of HCV RNA replication is not mediated through antagonizing LXRα–SREBP-1c activation. SfiI cells were treated with 50 μM arachidonic acid, EPA, or DHA alone or in the presence of the LXR agonist T0901317. Total RNA was harvested at 2 days posttreatment, and real-time RT-PCR analyses were performed to determine expression levels of SREBP-1c (A) and FAS (B). Northern blot analyses were performed to determine HCV RNA levels (C). Representative data (mean + SD) from three independent experiments are shown.
Similar articles
- Anti-ulcer agent teprenone inhibits hepatitis C virus replication: potential treatment for hepatitis C.
Ikeda M, Kawai Y, Mori K, Yano M, Abe K, Nishimura G, Dansako H, Ariumi Y, Wakita T, Yamamoto K, Kato N. Ikeda M, et al. Liver Int. 2011 Jul;31(6):871-80. doi: 10.1111/j.1478-3231.2011.02499.x. Epub 2011 Mar 13. Liver Int. 2011. PMID: 21645219 - Critical role of PA28gamma in hepatitis C virus-associated steatogenesis and hepatocarcinogenesis.
Moriishi K, Mochizuki R, Moriya K, Miyamoto H, Mori Y, Abe T, Murata S, Tanaka K, Miyamura T, Suzuki T, Koike K, Matsuura Y. Moriishi K, et al. Proc Natl Acad Sci U S A. 2007 Jan 30;104(5):1661-6. doi: 10.1073/pnas.0607312104. Epub 2007 Jan 18. Proc Natl Acad Sci U S A. 2007. PMID: 17234812 Free PMC article. - Establishment of a subgenomic replicon for bovine viral diarrhea virus in Huh-7 cells and modulation of interferon-regulated factor 3-mediated antiviral response.
Horscroft N, Bellows D, Ansari I, Lai VC, Dempsey S, Liang D, Donis R, Zhong W, Hong Z. Horscroft N, et al. J Virol. 2005 Mar;79(5):2788-96. doi: 10.1128/JVI.79.5.2788-2796.2005. J Virol. 2005. PMID: 15708997 Free PMC article. - Nonalcoholic steatosis and steatohepatitis. I. Molecular mechanism for polyunsaturated fatty acid regulation of gene transcription.
Clarke SD. Clarke SD. Am J Physiol Gastrointest Liver Physiol. 2001 Oct;281(4):G865-9. doi: 10.1152/ajpgi.2001.281.4.G865. Am J Physiol Gastrointest Liver Physiol. 2001. PMID: 11557505 Review. - [Replication of hepatitis C virus genome].
Kato N. Kato N. Uirusu. 2008 Dec;58(2):191-8. Uirusu. 2008. PMID: 19374197 Review. Japanese.
Cited by
- Positive-strand RNA virus replication organelles at a glance.
Stancheva VG, Sanyal S. Stancheva VG, et al. J Cell Sci. 2024 Sep 1;137(17):jcs262164. doi: 10.1242/jcs.262164. Epub 2024 Sep 10. J Cell Sci. 2024. PMID: 39254430 Free PMC article. Review. - Inhibition of sterol O-acyltransferase 1 blocks Zika virus infection in cell lines and cerebral organoids.
Schöbel A, Pinho Dos Reis V, Burkhard R, Hehner J, Schneider L, Schauflinger M, Vieyres G, Herker E. Schöbel A, et al. Commun Biol. 2024 Sep 5;7(1):1089. doi: 10.1038/s42003-024-06776-4. Commun Biol. 2024. PMID: 39237833 Free PMC article. - Metabolic Dysfunction-Associated Fatty Liver Disease and Chronic Viral Hepatitis: The Interlink.
Fernandez CJ, Alkhalifah M, Afsar H, Pappachan JM. Fernandez CJ, et al. Pathogens. 2024 Jan 10;13(1):68. doi: 10.3390/pathogens13010068. Pathogens. 2024. PMID: 38251375 Free PMC article. Review. - The Association between Statins and Liver Cancer Risk in Patients with Heart Failure: A Nationwide Population-Based Cohort Study.
Lu MC, Chen CC, Lu MY, Lin KJ, Chiu CC, Yang TY, Fang YA, Jian W, Chen MY, Hsu MH, Lai YH, Yang TL, Hao WR, Liu JC. Lu MC, et al. Cancers (Basel). 2023 May 29;15(11):2959. doi: 10.3390/cancers15112959. Cancers (Basel). 2023. PMID: 37296921 Free PMC article. - PEMT Mediates Hepatitis C Virus-Induced Steatosis, Explains Genotype-Specific Phenotypes and Supports Virus Replication.
Abomughaid M, Tay ESE, Pickford R, Malladi C, Read SA, Coorssen JR, Gloss BS, George J, Douglas MW. Abomughaid M, et al. Int J Mol Sci. 2023 May 15;24(10):8781. doi: 10.3390/ijms24108781. Int J Mol Sci. 2023. PMID: 37240132 Free PMC article.
References
- Bradley, D. W. (2000) Curr. Top. Microbiol. Immunol. 242, 1-23. - PubMed
- Alter, M. J., Margolis, H. S., Krawczynski, K., Judson, F. N., Mares, A., Alexander, W. J., Hu, P. Y., Miller, J. K., Gerber, M. A., Sampliner, R. E., et al. (1992) N. Engl. J. Med. 327, 1899-1905. - PubMed
- McCaffrey, A. P., Ohashi, K., Meuse, L., Shen, S., Lancaster, A. M., Lukavsky, P. J., Sarnow, P. & Kay, M. A. (2002) Mol. Ther. 5, 676-684. - PubMed
- Trowbridge, R. & Gowans, E. J. (1998) J. Viral Hepat. 5, 95-98. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources