Induction of Th1 type response by DNA vaccinations with N, M, and E genes against SARS-CoV in mice - PubMed (original) (raw)
Induction of Th1 type response by DNA vaccinations with N, M, and E genes against SARS-CoV in mice
Huali Jin et al. Biochem Biophys Res Commun. 2005.
Abstract
Vaccination against the SARS-CoV infection is an attractive means to control the spread of viruses in public. In this study, we employed a DNA vaccine technology with the levamisole, our newly discovered chemical adjuvant, to generate Th1 type of response. To avoid the enhancement antibody issue, genes encoding the nucleocapsid, membrane, and envelope protein of SARS-CoV were cloned and their expressions in mammalian cells were determined. After the intramuscular introduction into animals, we observed that the constructs of the E, M, and N genes could induce high levels of specific antibodies, T cell proliferations, IFN-gamma, DTH responses, and in vivo cytotoxic T cells activities specifically against SARS-CoV antigens. The highest immune responses were generated by the construct encoding the nucleocapsid protein. The results suggest that the N, M, and E genes could be used as the targets to prevent SARS-CoV infection in the DNA vaccine development.
Figures
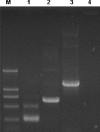
Fig. 1
Eukaryotic expression of DNA vaccines in HeLa cells. Total RNA was extracted from HeLa cells 12 h after transfected with pcD3d/E (lane 1), pcD3d/M (lane 2), pcD3d/N (lane 3), and pcD3d (lane 4). The RNAs were used to perform RT-PCR with specific primers, respectively. Positive band of each construct was observed in the transfected sample.
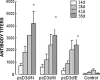
Fig. 2
Antibody responses to SARS-CoV mice (five per group) that were bled on day 14, 28, 42, and 56 after initial immunization. The mean titers with standard deviation (SD) of antibodies in five animals against CoV were evaluated by ELISA with 2 μg/ml of chemically killed SARS-CoV as antigen coated on each well in a 96-well plate and by using an absolute ratio of Post/Naïve serum at a cutoff of 2.1. *Indicates values that are statistically significant at a p value <0.05 by Student’s t test compared with the group of control vector.
Fig. 3
T cell proliferation with MTS colorimetric detection. Single suspension of splenocytes and lymphocytes of the immunized mice was isolated 2 weeks after the second immunization and stimulated in vitro either with the killed SARS-CoV for test groups or an unrelevant protein, BSA for negative control, or with Con A as the positive control. Splenocytes from a group of normal mice were stimulated with chemically killed SARS-CoV in vitro which served as the sham control. The stimulation index was derived from the value of test group divided by medium control group. (A) T cell proliferation responses from the spleen of the animals. (B) T cell proliferation responses from the spleen of the animals. *Indicates values that are statistically significant at a p value <0.05 by Student’s t test compared with all other groups.
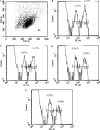
Fig. 4
In vivo CTL assay. To analyze SARS-CoV-specific cytotoxicity in vivo, a 1:1 mixture of 107 cells of each target cell population was injected i.v. into naïve BALB/c mice 7 days after the second injection with the DNA vaccine. After 4 h, the mice were killed and the splenocytes were analyzed for the presence of CFSEhigh and CFSElow target cell populations. The percentage of specific lysis was then determined as described in Materials and methods. (A) Representative histogram plot of lymph node cells obtained from the mice injected with SARS-CoV DNA vaccines 4 h after transfer of CFSE-labeled target cells. (B–E) The SARS-CoV-specific cytotoxic responses in the lymph nodes are shown from mice previously injected by the pcD3d vector control, pcD3d/E, pcD3d/M, and pcD3d/N constructs, respectively. Each datum has been repeated twice.
Fig. 5
Semi-quantitative RT-PCR assay for cytokine gene expression. Total RNA was isolated from spleen of the immunized groups or normal mice (3 for each group). The levels of the cytokines IL-2, IFN-γ, IL-4, and IL-10 were semi-quantitatively measured by a competitive RT-PCR with the addition of the 150 pg/ml of the pQRS competitor after the normalization of tested cDNA concentration to a constant amount of HPRT as described in Materials and methods.
Similar articles
- Protein transfer enhances cellular immune responses to DNA vaccination against SARS-CoV.
Kang Y, Chen A, Wang B, Zheng G. Kang Y, et al. Viral Immunol. 2009 Dec;22(6):417-22. doi: 10.1089/vim.2009.0048. Viral Immunol. 2009. PMID: 19951178 - Improvement in T helper 1-related immune responses in BALB/c mice immunized with an HIV-1 gag plasmid combined with a chimeric plasmid encoding interleukin-18 and flagellin.
Chen YL, Chen YS, Hung YC, Liu PJ, Tasi HY, Ni WF, Hseuh PT, Lin HH. Chen YL, et al. Microbiol Immunol. 2015 Aug;59(8):483-94. doi: 10.1111/1348-0421.12274. Microbiol Immunol. 2015. PMID: 26094825 - Levamisole promotes murine bone marrow derived dendritic cell activation and drives Th1 immune response in vitro and in vivo.
Fu Y, Wang T, Xiu L, Shi X, Bian Z, Zhang Y, Ruhan A, Wang X. Fu Y, et al. Int Immunopharmacol. 2016 Feb;31:57-65. doi: 10.1016/j.intimp.2015.12.015. Epub 2015 Dec 17. Int Immunopharmacol. 2016. PMID: 26706452 - CD8+ T Cells Responding to the Middle East Respiratory Syndrome Coronavirus Nucleocapsid Protein Delivered by Vaccinia Virus MVA in Mice.
Veit S, Jany S, Fux R, Sutter G, Volz A. Veit S, et al. Viruses. 2018 Dec 16;10(12):718. doi: 10.3390/v10120718. Viruses. 2018. PMID: 30558354 Free PMC article. - Ag85B of mycobacteria elicits effective CTL responses through activation of robust Th1 immunity as a novel adjuvant in DNA vaccine.
Takamura S, Matsuo K, Takebe Y, Yasutomi Y. Takamura S, et al. J Immunol. 2005 Aug 15;175(4):2541-7. doi: 10.4049/jimmunol.175.4.2541. J Immunol. 2005. PMID: 16081827
Cited by
- Nanotechnology in vaccine delivery.
Peek LJ, Middaugh CR, Berkland C. Peek LJ, et al. Adv Drug Deliv Rev. 2008 May 22;60(8):915-28. doi: 10.1016/j.addr.2007.05.017. Epub 2008 Feb 7. Adv Drug Deliv Rev. 2008. PMID: 18325628 Free PMC article. Review. - Construction of a bivalent DNA vaccine co-expressing S genes of transmissible gastroenteritis virus and porcine epidemic diarrhea virus delivered by attenuated Salmonella typhimurium.
Zhang Y, Zhang X, Liao X, Huang X, Cao S, Wen X, Wen Y, Wu R, Liu W. Zhang Y, et al. Virus Genes. 2016 Jun;52(3):354-64. doi: 10.1007/s11262-016-1316-z. Epub 2016 Mar 15. Virus Genes. 2016. PMID: 26980672 - Human memory T cell responses to SARS-CoV E protein.
Peng H, Yang LT, Li J, Lu ZQ, Wang LY, Koup RA, Bailer RT, Wu CY. Peng H, et al. Microbes Infect. 2006 Aug;8(9-10):2424-31. doi: 10.1016/j.micinf.2006.05.008. Epub 2006 Jun 30. Microbes Infect. 2006. PMID: 16844400 Free PMC article. - Intranasal immunization with plasmid DNA encoding spike protein of SARS-coronavirus/polyethylenimine nanoparticles elicits antigen-specific humoral and cellular immune responses.
Shim BS, Park SM, Quan JS, Jere D, Chu H, Song MK, Kim DW, Jang YS, Yang MS, Han SH, Park YH, Cho CS, Yun CH. Shim BS, et al. BMC Immunol. 2010 Dec 31;11:65. doi: 10.1186/1471-2172-11-65. BMC Immunol. 2010. PMID: 21194475 Free PMC article. - Persistent memory CD4+ and CD8+ T-cell responses in recovered severe acute respiratory syndrome (SARS) patients to SARS coronavirus M antigen.
Yang L, Peng H, Zhu Z, Li G, Huang Z, Zhao Z, Koup RA, Bailer RT, Wu C. Yang L, et al. J Gen Virol. 2007 Oct;88(Pt 10):2740-2748. doi: 10.1099/vir.0.82839-0. J Gen Virol. 2007. PMID: 17872527 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous