Recombinant modified vaccinia virus Ankara expressing the spike glycoprotein of severe acute respiratory syndrome coronavirus induces protective neutralizing antibodies primarily targeting the receptor binding region - PubMed (original) (raw)
. 2005 Mar;79(5):2678-88.
doi: 10.1128/JVI.79.5.2678-2688.2005.
Linqi Zhang, Chuan Qin, Lei Ba, Christopher E Yi, Fengwen Zhang, Qiang Wei, Tian He, Wenjie Yu, Jian Yu, Hong Gao, Xinming Tu, Agegnehu Gettie, Michael Farzan, Kwok-Yung Yuen, David D Ho
Affiliations
- PMID: 15708987
- PMCID: PMC548443
- DOI: 10.1128/JVI.79.5.2678-2688.2005
Recombinant modified vaccinia virus Ankara expressing the spike glycoprotein of severe acute respiratory syndrome coronavirus induces protective neutralizing antibodies primarily targeting the receptor binding region
Zhiwei Chen et al. J Virol. 2005 Mar.
Abstract
Immunization with a killed or inactivated viral vaccine provides significant protection in animals against challenge with certain corresponding pathogenic coronaviruses (CoVs). However, the promise of this approach in humans is hampered by serious concerns over the risk of leaking live severe acute respiratory syndrome (SARS) viruses. In this study, we generated a SARS vaccine candidate by using the live-attenuated modified vaccinia virus Ankara (MVA) as a vector. The full-length SARS-CoV envelope Spike (S) glycoprotein gene was introduced into the deletion III region of the MVA genome. The newly generated recombinant MVA, ADS-MVA, is replication incompetent in mammalian cells and highly immunogenic in terms of inducing potent neutralizing antibodies in mice, rabbits, and monkeys. After two intramuscular vaccinations with ADS-MVA alone, the 50% inhibitory concentration in serum was achieved with reciprocal sera dilutions of more than 1,000- to 10,000-fold in these animals. Using fragmented S genes as immunogens, we also mapped a neutralizing epitope in the region of N-terminal 400 to 600 amino acids of the S glycoprotein (S400-600), which overlaps with the angiotensin-converting enzyme 2 (ACE2) receptor-binding region (RBR; S318-510). Moreover, using a recombinant soluble RBR-Fc protein, we were able to absorb and remove the majority of the neutralizing antibodies despite observing that the full S protein tends to induce a broader spectrum of neutralizing activities in comparison with fragmented S proteins. Our data suggest that a major mechanism for neutralizing SARS-CoV likely occurs through blocking the interaction between virus and the cellular receptor ACE2. In addition, ADS-MVA induced potent immune responses which very likely protected Chinese rhesus monkeys from pathogenic SARS-CoV challenge.
Figures
FIG. 1.
Schematic representation of ADS-MVA construction (top and right panels) and the characterization of ADS-MVA by using avian cells (left bottom panel). The SARS-S gene was introduced, together with GFP gene, each under a separate promoter, into the Del III region of the MVA genome. (A) S glycoprotein was detected on CEF cells by using an anti-S polyclonal antibody (WH) in an immunofluorescence assay. (B) GFP coexpressed on the same cell population as seen in panel A.
FIG. 2.
Anti-SARS-CoV-specific neutralization antibody response in BALB/c mice after ADS-MVA vaccinations. Each mouse was designated with a number such as M260. The animals were immunized i.m. twice, 3 weeks apart, with 2 × 106 TCID50 (M260 and M262) or 2 × 107 TCID50 of ADS-MVA per mouse (M263, M264, and MM1 to MM4) or 2 × 107 TCID50 of ADC-MVA (M265 to M268). Two control mice (N1 and N2) only received saline under the same immunization schedule.
FIG. 3.
Anti-SARS-CoV specific neutralization antibody response in New Zealand White rabbits after ADS-MVA vaccinations. Each rabbit was designated with a number such as R524. The animals were immunized i.m. twice, 4 weeks apart, with 108 TCID50 of ADS-MVA per animal (R524 and R525) or 108 TCID50 of ADC-MVA (R520 and R521).
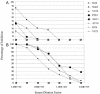
FIG. 4.
Anti-SARS-CoV-specific neutralization antibody response in Indian rhesus macaques after ADS-MVA vaccinations. Four animals (I624, N462, V876, and P283) were immunized i.m. twice, 4 weeks apart, with 1 × 108 and 3 × 108 TCID50 of ADS-MVA per animal, respectively. Another four macaques (AG11, AT14, I826, and V873) were treated the same but with ADC-MVA. (A) Neutralizing antibody responses after the first immunization. (B) Boosted neutralizing antibody responses after the second immunization.
FIG. 5.
Construction and expression of humanized fragmented S gene in the context of DNA vaccines. (A) Four S gene fragments (labeled A to D) were each constructed under cytomegalovirus (CMV) promoter in mammalian expression vector pcDNA3.1. (B) Fragmented S proteins of corresponding lengths were expressed in 293T cells in a Western blot analysis.
FIG. 6.
Anti-SARS-CoV-specific neutralization antibody response in New Zealand White rabbits after DNA vaccinations. Two animals per group were immunized twice, 4 weeks apart, using an in vivo electroporation technique with DNA vaccines S200 (R416 and R417), S400 (R418 and R419), S600 (R681 and R682), and S800 (R683 and R684). Another two rabbits were vaccinated with a DNA vaccine expressing HCVE1E2 gene (R518 and R519).
FIG. 7.
Absorption and removal of anti-SARS-CoV-specific neutralization antibodies of New Zealand White rabbits (A), BALB/c mice (B), and monkeys (C) by an ACE2 receptor-binding region and human Fc fusion protein (RBR-Fc). For rabbit and mouse sera, the neutralization antibody-RBR-Fc complex was removed by agarose beads that were precoated with specific anti-human Fc antibodies. The anti-human Fc antibodies do not cross-react with rabbit or mouse immunoglobulin but do cross-react with monkey samples. Therefore, for monkey sera, neutralization antibodies were removed by beads directly conjugated with RBR-Fc. In each experiment, the corresponding beads controls did not affect the activity of neutralization antibodies, as presented in groups of RBR-Fc(−). The experiment was carried out in triplicates, and the average values are presented. The error bars stand for the standard deviation.
Similar articles
- Identification of a critical neutralization determinant of severe acute respiratory syndrome (SARS)-associated coronavirus: importance for designing SARS vaccines.
He Y, Zhu Q, Liu S, Zhou Y, Yang B, Li J, Jiang S. He Y, et al. Virology. 2005 Mar 30;334(1):74-82. doi: 10.1016/j.virol.2005.01.034. Virology. 2005. PMID: 15749124 Free PMC article. - Heterologous MVA-S prime Ad5-S boost regimen induces high and persistent levels of neutralizing antibody response against SARS coronavirus.
Ba L, Yi CE, Zhang L, Ho DD, Chen Z. Ba L, et al. Appl Microbiol Biotechnol. 2007 Oct;76(5):1131-6. doi: 10.1007/s00253-007-1073-y. Epub 2007 Jun 21. Appl Microbiol Biotechnol. 2007. PMID: 17581748 Free PMC article. - Vaccine design for severe acute respiratory syndrome coronavirus.
He Y, Jiang S. He Y, et al. Viral Immunol. 2005;18(2):327-32. doi: 10.1089/vim.2005.18.327. Viral Immunol. 2005. PMID: 16035944 Review. - Severe acute respiratory syndrome vaccine development: experiences of vaccination against avian infectious bronchitis coronavirus.
Cavanagh D. Cavanagh D. Avian Pathol. 2003 Dec;32(6):567-82. doi: 10.1080/03079450310001621198. Avian Pathol. 2003. PMID: 14676007 Free PMC article. Review.
Cited by
- Transmission and Protection against Reinfection in the Ferret Model with the SARS-CoV-2 USA-WA1/2020 Reference Isolate.
Patel DR, Field CJ, Septer KM, Sim DG, Jones MJ, Heinly TA, Vanderford TH, McGraw EA, Sutton TC. Patel DR, et al. J Virol. 2021 Jun 10;95(13):e0223220. doi: 10.1128/JVI.02232-20. Epub 2021 Jun 10. J Virol. 2021. PMID: 33827954 Free PMC article. - T- and B-cell responses to multivalent prime-boost DNA and viral vectored vaccine combinations against hepatitis C virus in non-human primates.
Rollier CS, Verschoor EJ, Verstrepen BE, Drexhage JA, Paranhos-Baccala G, Liljeström P, Sutter G, Arribillaga L, Lasarte JJ, Bartosch B, Cosset FL, Inchauspe G, Heeney JL. Rollier CS, et al. Gene Ther. 2016 Oct;23(10):753-759. doi: 10.1038/gt.2016.55. Epub 2016 Jul 14. Gene Ther. 2016. PMID: 27416077 Free PMC article. - Emodin blocks the SARS coronavirus spike protein and angiotensin-converting enzyme 2 interaction.
Ho TY, Wu SL, Chen JC, Li CC, Hsiang CY. Ho TY, et al. Antiviral Res. 2007 May;74(2):92-101. doi: 10.1016/j.antiviral.2006.04.014. Epub 2006 May 15. Antiviral Res. 2007. PMID: 16730806 Free PMC article. - Severe acute respiratory syndrome (SARS) coronavirus: application of monoclonal antibodies and development of an effective vaccine.
Tsunetsugu-Yokota Y, Ohnishi K, Takemori T. Tsunetsugu-Yokota Y, et al. Rev Med Virol. 2006 Mar-Apr;16(2):117-31. doi: 10.1002/rmv.492. Rev Med Virol. 2006. PMID: 16518829 Free PMC article. Review.
References
- Altenburger, W., C. P. Suter, and J. Altenburger. 1989. Partial deletion of the human host range gene in the attenuated vaccinia virus MVA. Arch. Virol. 105:15-27. - PubMed
- Antoine, G., F. Scheiflinger, F. Dorner, and F. G. Falkner. 1998. The complete genomic sequence of the modified vaccinia Ankara strain: comparison with other orthopoxviruses. Virology 244:365-396. - PubMed
- Baron, S., J. Poast, D. Rizzo, E. McFarland, and E. Kieff. 2000. Electroporation of antibodies, DNA, and other macromolecules into cells: a highly efficient method. J. Immunol. Methods 242:115-126. - PubMed
- Booth, C. M., L. M. Matukas, G. A. Tomlinson, A. R. Rachlis, D. B. Rose, H. A. Dwosh, S. L. Walmsley, T. Mazzulli, M. Avendano, P. Derkach, I. E. Ephtimios, I. Kitai, B. D. Mederski, S. B. Shadowitz, W. L. Gold, L. A. Hawryluck, E. Rea, J. S. Chenkin, D. W. Cescon, S. M. Poutanen, and A. S. Detsky. 2003. Clinical features and short-term outcomes of 144 patients with SARS in the greater Toronto area. JAMA 289:2801-2809. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous