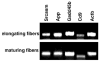Microarray analysis of fiber cell maturation in the lens - PubMed (original) (raw)
Microarray analysis of fiber cell maturation in the lens
Dmitry Ivanov et al. FEBS Lett. 2005.
Abstract
The mammalian lens consists of an aged core of quiescent cells enveloped by layers of mature fully elongated cells and younger, continuously elongating transcriptionally active cells. The fiber cell maturation is initiated when fiber cells cease to elongate. The process of maturation represents a radical switch from active elongation to a life-long quiescence and has not been studied previously. It may also include critical stages of preparation for the organelle removal and denucleation. In the present study, we used laser capture microdisection (LCM) microdissection and RNA amplification to compare global gene expression profiles of young elongating and mature, non-elongating fiber cells. Analysis of microarray data from three independent dye-swap experiments identified 65 differentially expressed genes (FDR<0.1) with greater than 2-fold change in expression levels. Microarray array results for a group of randomly selected genes were confirmed by quantitative RT-PCR. These microarray results provide clues to understanding the molecular pathways underlying lens development. The identified changes in the profile of gene expression reflected a shift in cell physiology characterizing the lens fiber maturation.
Figures
Fig. 1
Maturing and young fibers were discriminated in the TgN(GFPU5)Nagy mouse lenses using contrasting GFP labeling patterns (A). Mature fibers (Mt) were localized within the region of uniform GFP labeling, whereas young (Yg) fibers localized exclusively to the variegated region at the lens periphery. Paraffin sections of P5 lenses were microdissected by LCM using measurements performed on the contralateral eye (red and blue arrows correspond to the inner borders of the maturing and young fibers) (B). Cells cut from each of the two regions were collected. Bar is 50 mkm.
Fig. 2
The quality of the extracted RNA from elongating and maturing zones in the lens was determined by RT-PCR using primers for mouse Actb (β-actin), Gadd45b, App, Cd9, Srcasm.
Similar articles
- Comparative transcriptome analysis of epithelial and fiber cells in newborn mouse lenses with RNA sequencing.
Hoang TV, Kumar PK, Sutharzan S, Tsonis PA, Liang C, Robinson ML. Hoang TV, et al. Mol Vis. 2014 Nov 4;20:1491-517. eCollection 2014. Mol Vis. 2014. PMID: 25489224 Free PMC article. - Gene expression profiling in embryonic mouse lenses.
Xiao W, Liu W, Li Z, Liang D, Li L, White LD, Fox DA, Overbeek PA, Chen Q. Xiao W, et al. Mol Vis. 2006 Dec 26;12:1692-8. Mol Vis. 2006. PMID: 17213798 - The lens equator: a platform for molecular machinery that regulates the switch from cell proliferation to differentiation in the vertebrate lens.
Mochizuki T, Masai I. Mochizuki T, et al. Dev Growth Differ. 2014 Jun;56(5):387-401. doi: 10.1111/dgd.12128. Epub 2014 Apr 11. Dev Growth Differ. 2014. PMID: 24720470 Review. - The molecular mechanisms underlying lens fiber elongation.
Audette DS, Scheiblin DA, Duncan MK. Audette DS, et al. Exp Eye Res. 2017 Mar;156:41-49. doi: 10.1016/j.exer.2016.03.016. Epub 2016 Mar 23. Exp Eye Res. 2017. PMID: 27015931 Free PMC article. Review.
Cited by
- Transcriptional basis of the acclimation to high environmental temperature at the olfactory receptor organs of Drosophila melanogaster.
Riveron J, Boto T, Alcorta E. Riveron J, et al. BMC Genomics. 2013 Apr 17;14:259. doi: 10.1186/1471-2164-14-259. BMC Genomics. 2013. PMID: 23590196 Free PMC article. - RNA sequencing-based transcriptomic profiles of embryonic lens development for cataract gene discovery.
Anand D, Kakrana A, Siddam AD, Huang H, Saadi I, Lachke SA. Anand D, et al. Hum Genet. 2018 Dec;137(11-12):941-954. doi: 10.1007/s00439-018-1958-0. Epub 2018 Nov 11. Hum Genet. 2018. PMID: 30417254 Free PMC article. - RNA-binding proteins in eye development and disease: implication of conserved RNA granule components.
Dash S, Siddam AD, Barnum CE, Janga SC, Lachke SA. Dash S, et al. Wiley Interdiscip Rev RNA. 2016 Jul;7(4):527-57. doi: 10.1002/wrna.1355. Epub 2016 May 1. Wiley Interdiscip Rev RNA. 2016. PMID: 27133484 Free PMC article. Review. - A microarray study of MPP+-treated PC12 Cells: Mechanisms of toxicity (MOT) analysis using bioinformatics tools.
Xu Z, Patterson TA, Wren JD, Han T, Shi L, Duhart H, Ali SF, Slikker W Jr. Xu Z, et al. BMC Bioinformatics. 2005 Jul 15;6 Suppl 2(Suppl 2):S8. doi: 10.1186/1471-2105-6-S2-S8. BMC Bioinformatics. 2005. PMID: 16026605 Free PMC article. - Apoptosis gene profiling reveals spatio-temporal regulated expression of the p53/Mdm2 pathway during lens development.
Geatrell JC, Gan PM, Mansergh FC, Kisiswa L, Jarrin M, Williams LA, Evans MJ, Boulton ME, Wride MA. Geatrell JC, et al. Exp Eye Res. 2009 Jun;88(6):1137-51. doi: 10.1016/j.exer.2009.01.020. Epub 2009 Feb 11. Exp Eye Res. 2009. PMID: 19450442 Free PMC article.
References
- Bhat SP. The ocular lens epithelium. Biosci Rep. 2001;21:537–600. Review. - PubMed
- Piatigorsky J.1981Lens differentiation in vertebrates. A review of cellular and molecular features Differentiation 19134187Review. - PubMed
- Beebe DC, Compart PJ, Johnson MC, Feagans DE, Feinberg RN. The mechanism of cell elongation during lens fiber cell differentiation. Dev Biol. 1982;92:54–63. - PubMed
- Bassnett S, Beebe DC. Coincident loss of mitochondria and nuclei during lens fiber cell differentiation. Dev Dyn. 1992;194:85–93. - PubMed
- Bassnett S. Lens organelle degradation. Exp Eye Res. 2002;74:1–6. Review. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases