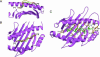Crystal structure of HLA-G: a nonclassical MHC class I molecule expressed at the fetal-maternal interface - PubMed (original) (raw)
Crystal structure of HLA-G: a nonclassical MHC class I molecule expressed at the fetal-maternal interface
Craig S Clements et al. Proc Natl Acad Sci U S A. 2005.
Abstract
HLA-G is a nonclassical major histocompatibility complex class I (MHC-I) molecule that is primarily expressed at the fetal-maternal interface, where it is thought to play a role in protecting the fetus from the maternal immune response. HLA-G binds a limited repertoire of peptides and interacts with the inhibitory leukocyte Ig-like receptors LIR-1 and LIR-2 and possibly with certain natural killer cell receptors. To gain further insights into HLA-G function, we determined the 1.9-A structure of a monomeric HLA-G complexed to a natural endogenous peptide ligand from histone H2A (RIIPRHLQL). An extensive network of contacts between the peptide and the antigen-binding cleft reveal a constrained mode of binding reminiscent of the nonclassical HLA-E molecule, thereby providing a structural basis for the limited peptide repertoire of HLA-G. The alpha3 domain of HLA-G, a candidate binding site for the LIR-1 and -2 inhibitory receptors, is structurally distinct from the alpha3 domains of classical MHC-I molecules, providing a rationale for the observed affinity differences for these ligands. The structural data suggest a head-to-tail mode of dimerization, mediated by an intermolecular disulfide bond, that is consistent with the observation of HLA-G dimers on the cell surface.
Figures
Fig. 1.
Overview of the structure of HLA-G. (A) The hc is shown in purple, β2M in blue, and the peptide in green. The position of the Cys-42 → Ser mutation is labeled. (B) Conformation of bound peptide. The corresponding 2_F_o - _F_c electron density is shown in mesh format. The peptide is in ball-and-stick format with each amino acid labeled.
Fig. 2.
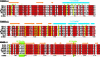
Sequence alignment of human HLA-G with HLA-E, -A2, -B44, and -CW3 as well as mouse Qa-2. The sequence alignment is divided into the three domains: α1, α2, and α3. Residues highlighted with red have 100% identity. Residues highlighted in yellow are conservatively substituted. The secondary structure of HLA-G is illustrated directly above the sequence alignment. Orange arrows indicate β-strands, and blue cylinders indicate α-helices. Residues directly interacting with the peptide are marked with a large green star, and residues interacting only via a water molecule are marked with a hollow circle (see Table 3). The loop in domain α3 involved in LIR-1/2 (ILT-2/4) binding is highlighted with a green box.
Fig. 3.
Comparison of MHC-I bound peptides. The conformations of peptides bound by HLA-G (purple), HLA-E (48) (red), HLA-A2 (49) (yellow), and Qa-2 (39) (green) are shown in a side view (A), top view (B), and a view showing the main chain interactions between the epitope and the HLA-G hc (C). Polar interactions are depicted only (dashed lines); water molecules are shown in blue spheres.
Fig. 4.
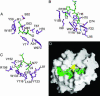
Pocket-mediated interactions of HLA-G. HLA-G residues are shown in purple, peptide residues are shown in green, polar contacts are depicted as dashed lines, and water molecules are shown as blue spheres. (A) Pockets A and B mediating interactions with P1-Arg and P2-Ile, respectively. (B) Pockets C and D mediating interactions with P6-His and P2-Pro, respectively. (C) Pockets E and F mediating interactions with P7-Leu and P9-Leu, respectively. (D) Overview of the hydrophobic nature of the HLA-G binding cleft. Hydrophobic regions of the cleft that bind peptide are shown in green with the important anchor residue His-70 colored yellow. The pockets are labeled A–F, and the peptide is depicted as a ball-and-stick structure.
Fig. 5.
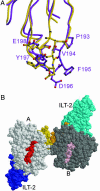
HLA-G may bind to LIR-1 (ILT-2) with enhanced affinity and undergo disulfide-bond-mediated dimerization. (A) Comparative analysis of the candidate ILT-2 binding site on the α3 domain of HLA-G (purple) and the equivalent binding site on HLA-A2 (yellow). (B) Model of the HLA-G dimer, looking down onto the antigen-binding cleft; peptides are in red (A protomer, light gray) and pink (B protomer, dark gray). The dimerization site is mediated via an intermolecular Cys-42–Cys-42 disulfide, indicating that the protomers within the dimer cannot adopt a side-by-side arrangement. However, a head-to-tail HLA-G dimer would satisfy the requirements for a Cys-42-mediated dimer, in which the loops containing the Cys-42 would clasp onto each other. In this plausible configuration, intermolecular interactions mediated via the β2M domains are possible. LIR-1 has been solved in complex with HLA-A2 (Protein Data Bank ID code 1P7Q), in which the LIR-1 binding site onto the α3 domain of the hc was mapped. Given the similarities between HLA-G and HLA-A2, the molecular organization of the HLA-G/ILT2 complex can be postulated in this HLA-G dimeric arrangement (ILT2 in blue and cyan).
Similar articles
- Structural studies on HLA-G: implications for ligand and receptor binding.
Clements CS, Kjer-Nielsen L, McCluskey J, Rossjohn J. Clements CS, et al. Hum Immunol. 2007 Apr;68(4):220-6. doi: 10.1016/j.humimm.2006.09.003. Epub 2006 Oct 27. Hum Immunol. 2007. PMID: 17400055 Review. - Human inhibitory receptors Ig-like transcript 2 (ILT2) and ILT4 compete with CD8 for MHC class I binding and bind preferentially to HLA-G.
Shiroishi M, Tsumoto K, Amano K, Shirakihara Y, Colonna M, Braud VM, Allan DS, Makadzange A, Rowland-Jones S, Willcox B, Jones EY, van der Merwe PA, Kumagai I, Maenaka K. Shiroishi M, et al. Proc Natl Acad Sci U S A. 2003 Jul 22;100(15):8856-61. doi: 10.1073/pnas.1431057100. Epub 2003 Jul 9. Proc Natl Acad Sci U S A. 2003. PMID: 12853576 Free PMC article. - Natural killer cell recognition of HLA class I molecules.
Brooks AG, Boyington JC, Sun PD. Brooks AG, et al. Rev Immunogenet. 2000;2(3):433-48. Rev Immunogenet. 2000. PMID: 11256749 Review. - Crystal structure of HLA-A2 bound to LIR-1, a host and viral major histocompatibility complex receptor.
Willcox BE, Thomas LM, Bjorkman PJ. Willcox BE, et al. Nat Immunol. 2003 Sep;4(9):913-9. doi: 10.1038/ni961. Epub 2003 Aug 3. Nat Immunol. 2003. PMID: 12897781 - Immune modulation of HLA-G dimer in maternal-fetal interface.
Kuroki K, Maenaka K. Kuroki K, et al. Eur J Immunol. 2007 Jul;37(7):1727-9. doi: 10.1002/eji.200737515. Eur J Immunol. 2007. PMID: 17587197 Free PMC article. Review.
Cited by
- HLA-G Neo-Expression on Tumors.
Loustau M, Anna F, Dréan R, Lecomte M, Langlade-Demoyen P, Caumartin J. Loustau M, et al. Front Immunol. 2020 Aug 14;11:1685. doi: 10.3389/fimmu.2020.01685. eCollection 2020. Front Immunol. 2020. PMID: 32922387 Free PMC article. Review. - Emerging topics and new perspectives on HLA-G.
Fainardi E, Castellazzi M, Stignani M, Morandi F, Sana G, Gonzalez R, Pistoia V, Baricordi OR, Sokal E, Peña J. Fainardi E, et al. Cell Mol Life Sci. 2011 Feb;68(3):433-51. doi: 10.1007/s00018-010-0584-3. Epub 2010 Nov 16. Cell Mol Life Sci. 2011. PMID: 21080027 Free PMC article. Review. - Preimplantation embryo development (Ped) gene copy number varies from 0 to 85 in a population of wild mice identified as Mus musculus domesticus.
Byrne MJ, Jones GS, Warner CM. Byrne MJ, et al. Mamm Genome. 2007 Nov;18(11):767-78. doi: 10.1007/s00335-007-9067-8. Epub 2007 Nov 8. Mamm Genome. 2007. PMID: 17990033 - Non-classical transcriptional regulation of HLA-G: an update.
Moreau P, Flajollet S, Carosella ED. Moreau P, et al. J Cell Mol Med. 2009 Sep;13(9B):2973-89. doi: 10.1111/j.1582-4934.2009.00800.x. Epub 2009 Jun 5. J Cell Mol Med. 2009. PMID: 19508383 Free PMC article. Review. - Nitric oxide produces HLA-G nitration and induces metalloprotease-dependent shedding creating a tolerogenic milieu.
Díaz-Lagares A, Alegre E, LeMaoult J, Carosella ED, González A. Díaz-Lagares A, et al. Immunology. 2009 Mar;126(3):436-45. doi: 10.1111/j.1365-2567.2008.02911.x. Epub 2008 Sep 1. Immunology. 2009. PMID: 18764882 Free PMC article.
References
- Margulies, D. & McCluskey, J. (2003) in Fundamental Immunology, ed. Paul, W. (Lippincott, Philadelphia).
- Bjorkman, P. J. & Parham, P. (1990) Annu. Rev. Biochem. 59, 253-288. - PubMed
- O'Callaghan, C. A. & Bell, J. I. (1998) Immunol. Rev. 163, 129-138. - PubMed
- Gao, G. F., Willcox, B. E., Wyer, J. R., Boulter, J. M., O'Callaghan, C. A., Maenaka, K., Stuart, D. I., Jones, E. Y., Van Der Merwe, P. A., Bell, J. I. & Jakobsen, B. K. (2000) J. Biol. Chem. 275, 15232-15238. - PubMed
- Rammensee, H. G., Falk, K. & Rotzschke, O. (1993) Annu. Rev. Immunol. 11, 213-244. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials