Signal transducer and activator of transcription (STAT) signalling and T-cell lymphomas - PubMed (original) (raw)
Review
Signal transducer and activator of transcription (STAT) signalling and T-cell lymphomas
Tracey J Mitchell et al. Immunology. 2005 Mar.
Abstract
Interaction of cytokines with their cognate receptors leads to the activation of latent transcription factors - the signal transducers and activators of transcription (STAT) proteins - whose biological activities ultimately regulate many critical aspects of cell growth, survival and differentiation. Dysregulation of the JAK-STAT pathway is frequently observed in many primary human tumours, reflecting the importance of this pathway in the maintenance of cellular integrity. Here we review the current progress in STAT structure and function, and the contribution of STAT signalling to the pathogenesis of T-cell lymphomas.
Figures
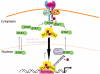
Figure 1
The JAK-signal transducer and activator of transcription (JAK-STAT) signalling pathway. Ligand-induced activation of latent cytoplasmic JAK kinases leads to the docking and subsequent tyrosine phosphorylation of latent monomeric or N-domain-mediated dimeric STAT proteins, facilitating the formation of active SH2-domain-mediated homodimers of STAT proteins. The tyrosine-phosphorylated STAT dimers are actively transported to the nucleus using metabolic energy and the importin α/β and RanGDP complex. Once inside the nucleus, the active STAT dimers bind to the promoters of genes containing the consensus recognition motif (GAS motif-ttcnnngaa) and activate transcription of these genes. STATs can bind DNA as dimers or as N-domain-mediated tetramers. The active STAT protein is only released from DNA upon dephosphorylation by nuclear phosphatases, following cytokine withdrawal, and the inactivated protein is then actively exported out of the nucleus to the cytoplasm by the exportin Crm-1/RanGTP complex. Monomeric STAT proteins can also passively shuttle between the cytoplasm and the nucleus via carrier-free diffusion through nuclear pores facilitated by the interaction with nucleoporins.
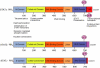
Figure 2
Modular structure of signal transducer and activator of transcription (STAT) proteins. All STAT proteins share a common molecular topology and are organized into distinct functional domains. The NH2 terminus (N-domain) is involved in protein–protein interactions between adjacent STAT dimers on DNA, facilitating the formation of STAT tetramers. It is also involved in the formation of dimers between non-phosphorylated STAT monomers, which is important for receptor-mediated activation and nuclear translocation of certain STAT proteins. Interactions with STAT cofactors, which positively or negatively modulate their transcriptional activity, occurs via the N-domain, the adjacent coiled-coil domain and the carboxy-terminal transactivation domain (TAD). The conserved serine residue (p-S), which is phosphorylated upon cytokine stimulation and is important for maximal transcriptional activation, is located within the transactivation domain. In addition to the full-length form (STATα), STAT proteins can also be present as C-terminally truncated forms generated by alternative splicing (STATβ) or by proteolytic processing (STATγ). The C-termini of STATβ proteins differs from the corresponding STATα protein not only by being truncated but also by having additional amino acids inserted as a result of the splicing event.
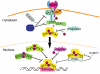
Figure 3
Negative regulation of signal transducer and activator of transcription (STAT) signalling. Cytokine-induced STAT activation can be inhibited by SOCS proteins, the gene expression of which is regulated by STAT proteins, thus fulfilling a negative feedback loop. SOCS proteins inhibit STAT activation either by inhibition of the activating JAKs or by competition with STATs for receptor binding. Activated STAT proteins can be dephosphorylated by cytoplasmic and/or nuclear phosphatases. C-terminally truncated STAT proteins, STATβ and STATγ, behave as dominant-negative proteins to functionally compete with their full-length counterparts to alter or inhibit gene expression, respectively. Protein inhibitors of activated STAT proteins (PIAS) interact with STAT proteins to inhibit their DNA binding and/or potentially facilitate their covalent modification by sumoylation and subsequent degradation.
Similar articles
- JAK/STAT-dependent gene regulation by cytokines.
Hebenstreit D, Horejs-Hoeck J, Duschl A. Hebenstreit D, et al. Drug News Perspect. 2005 May;18(4):243-9. doi: 10.1358/dnp.2005.18.4.908658. Drug News Perspect. 2005. PMID: 16034480 Review. - STAT proteins as novel targets for cancer drug discovery.
Turkson J. Turkson J. Expert Opin Ther Targets. 2004 Oct;8(5):409-22. doi: 10.1517/14728222.8.5.409. Expert Opin Ther Targets. 2004. PMID: 15469392 Review. - Cytokines and STAT signaling.
Schindler C, Strehlow I. Schindler C, et al. Adv Pharmacol. 2000;47:113-74. doi: 10.1016/s1054-3589(08)60111-8. Adv Pharmacol. 2000. PMID: 10582086 Review. No abstract available. - Signal transducer and activator of transcription proteins in leukemias.
Benekli M, Baer MR, Baumann H, Wetzler M. Benekli M, et al. Blood. 2003 Apr 15;101(8):2940-54. doi: 10.1182/blood-2002-04-1204. Epub 2002 Dec 12. Blood. 2003. PMID: 12480704 Review. - Prolactin signal transduction mechanisms in the mammary gland: the role of the Jak/Stat pathway.
Watson CJ, Burdon TG. Watson CJ, et al. Rev Reprod. 1996 Jan;1(1):1-5. doi: 10.1530/ror.0.0010001. Rev Reprod. 1996. PMID: 9414431 Review.
Cited by
- Oligonucleotide-Based Therapeutics for STAT3 Targeting in Cancer-Drug Carriers Matter.
Molenda S, Sikorska A, Florczak A, Lorenc P, Dams-Kozlowska H. Molenda S, et al. Cancers (Basel). 2023 Nov 29;15(23):5647. doi: 10.3390/cancers15235647. Cancers (Basel). 2023. PMID: 38067351 Free PMC article. Review. - Exploring the molecular mechanism of ginseng against anthracycline-induced cardiotoxicity based on network pharmacology, molecular docking and molecular dynamics simulation.
Xie L, Liu H, Zhang K, Pan Y, Chen M, Xue X, Wan G. Xie L, et al. Hereditas. 2024 Sep 6;161(1):31. doi: 10.1186/s41065-024-00334-y. Hereditas. 2024. PMID: 39243097 Free PMC article. - Role of Mitogen Activated Protein Kinase Signaling in Parkinson's Disease.
Bohush A, Niewiadomska G, Filipek A. Bohush A, et al. Int J Mol Sci. 2018 Sep 29;19(10):2973. doi: 10.3390/ijms19102973. Int J Mol Sci. 2018. PMID: 30274251 Free PMC article. Review. - microRNA-141-3p-containing small extracellular vesicles derived from epithelial ovarian cancer cells promote endothelial cell angiogenesis through activating the JAK/STAT3 and NF-κB signaling pathways.
Masoumi-Dehghi S, Babashah S, Sadeghizadeh M. Masoumi-Dehghi S, et al. J Cell Commun Signal. 2020 Jun;14(2):233-244. doi: 10.1007/s12079-020-00548-5. Epub 2020 Feb 7. J Cell Commun Signal. 2020. PMID: 32034654 Free PMC article. - STAT3 inhibitors: finding a home in lymphoma and leukemia.
Munoz J, Dhillon N, Janku F, Watowich SS, Hong DS. Munoz J, et al. Oncologist. 2014 May;19(5):536-44. doi: 10.1634/theoncologist.2013-0407. Epub 2014 Apr 4. Oncologist. 2014. PMID: 24705981 Free PMC article. Review.
References
- Darnell JE, Jr, Kerr IM, Stark GR. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science. 1994;264:1415. - PubMed
- Gadina M, Hilton D, Johnston JA, Morinobu A, Lighvani A, Zhou YJ, Visconti R, O'Shea JJ. Signaling by type I and II cytokine receptors: ten years after. Curr Opin Immunol. 2001;13:363. - PubMed
- Bowman T, Garcia R, Turkson J, Jove R. STATs in oncogenesis. Oncogene. 2000;19:2474. - PubMed
- Calo V, Migliavacca M, Bazan V, Macaluso M, Buscemi M, Gebbia N, Russo A. STAT proteins: from normal control of cellular events to tumorigenesis. J Cell Physiol. 2003;197:157. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous