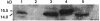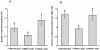A Vibrio vulnificus type IV pilin contributes to biofilm formation, adherence to epithelial cells, and virulence - PubMed (original) (raw)
A Vibrio vulnificus type IV pilin contributes to biofilm formation, adherence to epithelial cells, and virulence
Rohinee N Paranjpye et al. Infect Immun. 2005 Mar.
Abstract
Vibrio vulnificus expresses a multitude of cell-associated and secreted factors that potentially contribute to pathogenicity, although the specific roles of most of these factors have been difficult to define. Previously we have shown that a mutation in pilD (originally designated vvpD), which encodes a type IV prepilin peptidase/N-methyltransferase, abolishes expression of surface pili, suggesting that they belong to the type IV class. In addition, a pilD mutant exhibits reduced adherence to HEp-2 cells, a block in secretion of several exoenzymes that follow the type II secretion pathway, and decreased virulence. In this study, we have cloned and characterized a V. vulnificus type IV pilin (PilA) that shares extensive homology to group A type IV pilins expressed by many pathogens, including Vibrio cholerae (PilA), Pseudomonas aeruginosa (PilA), and Aeromonas hydrophila (TapA). The V. vulnificus pilA gene is part of an operon and is clustered with three other pilus biogenesis genes, pilBCD. Inactivation of pilA reduces the ability of V. vulnificus to form biofilms and significantly decreases adherence to HEp-2 cells and virulence in iron dextran-treated mice. Southern blot analysis demonstrates the widespread presence of both pilA and pilD in clinical as well as environmental strains of V. vulnificus.
Figures
FIG. 1.
Genetic organization of the type IV pilus biogenesis gene cluster of V. vulnificus. (A) Relevant restriction sites used for cloning constructs and probes. (B) Comparison of the type IV pilus biogenesis pil genes of V. vulnificus with those of V. cholerae, A. hydrophila, A. salmonicida, and P. aeruginosa.
FIG. 2.
(A) Comparison of the amino acid sequence of V. vulnificus PilA with the homologous type IV pilins of V. cholerae (Vc PilA), A. hydrophila (Ah TapA), P. aeruginosa (Pa PilA), and V. cholerae (Vc MshA). Conserved residues around the cleavage site are shaded. The remainder of the sequence is less homologous. The inverted triangle indicates the consensus cleavage site for group A, type IV pilins. (B) Comparison of the amino acid sequence of V. vulnificus PilB with homologs from V. cholerae (Vc PilB), A. hydrophila (Ah TapB), and P. aeruginosa (Pa PilB). The location of the Walker box motif is shaded. Both alignments were generated by using the Pileup program of the Genetics Computer Group (Madison, Wis.).
FIG. 3.
Expression of PilA in V. vulnificus strains grown in TCG medium at 30°C. Whole-cell lysates were analyzed by Western blotting with anti-PilA antiserum. Lane 1, C7184 (wild-type); lane 2, C7184AΩ (pilA mutant); lane 3, C7184AΩ (pilA +); lane 4, C7184DΩ (pilA +); lane 5, PilA (purified).
FIG. 4.
Detection of pilA, pilB, pilC, and pilD transcripts by RT-PCR in V. vulnificus C7184 (wild type) (lanes 1 to 4) and C7184AΩ (pilA mutant) strains (lanes 5 to 8). The corresponding control RT reactions in which the addition of Superscript II RNase H− reverse transcriptase was omitted are shown in lanes 9 to 16.
FIG. 5.
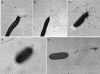
Transmission electron micrographs of V. vulnificus C7184 strains showing surface pili (indicated by arrows). Cells were negatively stained with 2% uranyl acetate on Butyar-coated grids. (A) C7184; (B) C7184AΩ; (C) C7184AΩ(pRP383); (D) P. aeruginosa PAK-NP; (E) P. aeruginosa PAK-NP(pRP383). Magnification, ×15,500.
FIG. 6.
Comparison of adherence of V. vulnificus strains to epithelial cells (HEp-2). (A) Adherence was determined with V. vulnificus strains labeled with [methyl-3H]thymidine. (B) One hundred epithelial cells were counted for each strain. Values represent the mean number ± standard deviation of adherent bacteria per coverslip (A) or the number of adherent bacteria per epithelial cell (B). Values denoted by different letters are significantly different when compared by a one-way ANOVA and Fisher's PLSD at P ≤ 0.05.
FIG. 7.
Southern blot analysis of clinical and environmental isolates of V. vulnificus, using a probe encompassing the pilA gene (see Materials and Methods). I, clinical isolates; S, shellfish isolates associated with illness; O, oyster isolates not associated with illness. M, strain MO6-24; P, PAC1; C, C7184.
Similar articles
- An Aeromonas salmonicida type IV pilin is required for virulence in rainbow trout Oncorhynchus mykiss.
Masada CL, LaPatra SE, Morton AW, Strom MS. Masada CL, et al. Dis Aquat Organ. 2002 Aug 15;51(1):13-25. doi: 10.3354/dao051013. Dis Aquat Organ. 2002. PMID: 12240967 - The type IV leader peptidase/N-methyltransferase of Vibrio vulnificus controls factors required for adherence to HEp-2 cells and virulence in iron-overloaded mice.
Paranjpye RN, Lara JC, Pepe JC, Pepe CM, Strom MS. Paranjpye RN, et al. Infect Immun. 1998 Dec;66(12):5659-68. doi: 10.1128/IAI.66.12.5659-5668.1998. Infect Immun. 1998. PMID: 9826339 Free PMC article. - Role of type IV pilins in persistence of Vibrio vulnificus in Crassostrea virginica oysters.
Paranjpye RN, Johnson AB, Baxter AE, Strom MS. Paranjpye RN, et al. Appl Environ Microbiol. 2007 Aug;73(15):5041-4. doi: 10.1128/AEM.00641-07. Epub 2007 Jun 8. Appl Environ Microbiol. 2007. PMID: 17557854 Free PMC article. - Type II secretion and type IV pili of Francisella.
Forsberg A, Guina T. Forsberg A, et al. Ann N Y Acad Sci. 2007 Jun;1105:187-201. doi: 10.1196/annals.1409.016. Epub 2007 Apr 13. Ann N Y Acad Sci. 2007. PMID: 17435117 Review. - Molecular Pathogenesis of Vibrio vulnificus.
Gulig PA, Bourdage KL, Starks AM. Gulig PA, et al. J Microbiol. 2005 Feb;43 Spec No:118-31. J Microbiol. 2005. PMID: 15765065 Review.
Cited by
- Insight Into the Virulence Related Secretion Systems, Fimbriae, and Toxins in O2:K1 Escherichia coli Isolated From Bovine Mastitis.
Sun M, Gao X, Zhao K, Ma J, Yao H, Pan Z. Sun M, et al. Front Vet Sci. 2021 Feb 11;8:622725. doi: 10.3389/fvets.2021.622725. eCollection 2021. Front Vet Sci. 2021. PMID: 33644149 Free PMC article. - Roles of putative type II secretion and type IV pilus systems in the virulence of uropathogenic Escherichia coli.
Kulkarni R, Dhakal BK, Slechta ES, Kurtz Z, Mulvey MA, Thanassi DG. Kulkarni R, et al. PLoS One. 2009;4(3):e4752. doi: 10.1371/journal.pone.0004752. Epub 2009 Mar 9. PLoS One. 2009. PMID: 19270734 Free PMC article. - Environmental Factors Influencing Occurrence of Vibrio parahaemolyticus and Vibrio vulnificus.
Brumfield KD, Chen AJ, Gangwar M, Usmani M, Hasan NA, Jutla AS, Huq A, Colwell RR. Brumfield KD, et al. Appl Environ Microbiol. 2023 Jun 28;89(6):e0030723. doi: 10.1128/aem.00307-23. Epub 2023 May 24. Appl Environ Microbiol. 2023. PMID: 37222620 Free PMC article. - Genotypic Diversity and Population Structure of Vibrio vulnificus Strains Isolated in Taiwan and Korea as Determined by Multilocus Sequence Typing.
Kim HJ, Cho JC. Kim HJ, et al. PLoS One. 2015 Nov 23;10(11):e0142657. doi: 10.1371/journal.pone.0142657. eCollection 2015. PLoS One. 2015. PMID: 26599487 Free PMC article. - The HlyU protein is a positive regulator of rtxA1, a gene responsible for cytotoxicity and virulence in the human pathogen Vibrio vulnificus.
Liu M, Alice AF, Naka H, Crosa JH. Liu M, et al. Infect Immun. 2007 Jul;75(7):3282-9. doi: 10.1128/IAI.00045-07. Epub 2007 Apr 16. Infect Immun. 2007. PMID: 17438022 Free PMC article.
References
- Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. - PubMed
- Bieber, D., S. W. Ramer, C. Y. Wu, W. J. Murray, T. Tobe, R. Fernandez, and G. K. Schoolnik. 1998. Type IV pili, transient bacterial aggregates, and virulence of enteropathogenic Escherichia coli. Science 280:2114-2118. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials