The C-terminal half of TSG101 blocks Rous sarcoma virus budding and sequesters Gag into unique nonendosomal structures - PubMed (original) (raw)
The C-terminal half of TSG101 blocks Rous sarcoma virus budding and sequesters Gag into unique nonendosomal structures
Marc C Johnson et al. J Virol. 2005 Mar.
Abstract
Retroviral late domains (L domains) are short amino acid sequences in the Gag protein that facilitate the process of budding. L domains act by recruiting the ESCRT complexes, which normally function in the formation of multivesicular bodies. The PTAP late domain of human immunodeficiency virus (HIV) is believed to specifically recruit this machinery by binding the ESCRT protein TSG101. It was recently demonstrated that expression of a C-terminal fragment of TSG101 (TSG-3') blocked the budding of both PTAP-dependent and PPPY-dependent retroviruses. We show here that TSG-3' expression leads to the formation of large spherical entities that we call TICS (TSG-3'-induced cellular structures) in the cytoplasm. Rous sarcoma virus (RSV) and murine leukemia virus (MLV) Gag proteins are selectively recruited to these structures, but HIV type 1 Gag is completely excluded. Experiments with various HIV and RSV vector constructs as well as HIV and RSV chimeras suggest that recruitment to the TICS is late domain independent and does not involve recognition of any single amino acid sequence. TICS appear to have no limiting membrane and do not colocalize with markers for any membranous cellular compartment. Wild-type TSG101 is also recruited to TICS, but most other ESCRT proteins are excluded. These structures are similar in nature to aggresomes, colocalize with the aggresome marker GFP-250, and are highly enriched in ubiquitin but in other ways do not fully meet the description of aggresomes. We conclude that the block to retroviral budding by TSG-3' may be the result of its sequestration of Gag, depletion of free TSG101, or depletion of free ubiquitin.
Figures
FIG. 1.
Vectors used in this study. HRH-GFP and RHR-GFP are chimeras of RSV Gag and HIV Gag. Light-gray domains represent fluorescent tags. Dark-gray boxes represent HIV sequence. A full description of all vectors is in Materials and Methods. UEV, ubiquitin E2 variant domain; PRD, proline-rich domain; coil, coiled-coil domain.
FIG. 2.
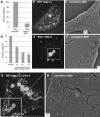
RSV Gag sequestration by TSG-3′. (A) Budding efficiency of RSV Gag-GFP(ΔL). Gag-GFP was normalized to 100%. (B, E, G) Fluorescence-merged Z sections of cells transfected with a mixture of DNAs at the ratios indicated. (B) Nine-to-one ratio of RSV Gag(ΔL) to RSV Gag(ΔL)-GFP; (E) 9:1:10 DNA ratio of RSV Gag to RSV Gag-GFP to TSG-3′; (G) 9:1:10 ratio of RSV Gag(ΔL) to RSV Gag(ΔL)-GFP to TSG-3′. (C, F, H) Corresponding SEM images of the surfaces of the same individual cells as shown in panels B, E, and G, respectively. (D) Budding efficiency of Gag in cells cotransfected with RSV Gag and TSG-3′ at the ratios indicated. Percentages released are expressed relative to the release of parallel cells transfected with an equal amount of Gag.
FIG. 3.
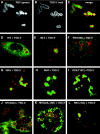
Recruitment of Gag proteins by TSG-3′. (A-C) Cells transfected with a 1:1 ratio of RSV Gag-GFP to TSG-3′. (A) RSV Gag-GFP. (B) Immunofluorescence visualization of the HA tag of TSG-3′. (C) Merge of panels A and B. (D-L) Cells cotransfected with a 1:1 ratio of TSG-3′ to individual GFP or YFP-tagged Gag vectors. Each image is a merge of the TSG-3′ immunofluorescence image (red) and the fluorescent Gag image (green). (D) HIV Gag-GFP. (E) MLV Gag-YFP. (F) RSV Gag(SM)-GFP. (G) HRH-GFP. (H) RHR-GFP. (I) RSV Gag(T10C)-GFP. (J) HIV Gag(G2A)-GFP. (K) HIV Gag(G2A,ΔGH)-YFP. (L) RSV Gag(ΔNC)-GFP.
FIG. 4.
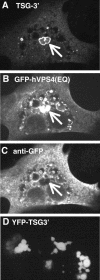
Antibody staining of TSG-3′ compartments. (A-C) Cell transfected with a 1:1 ratio of TSG-3′ to GFP-hVPS4(EQ). (A) Immunofluorescent stain against the HA tag of TSG-3′. (B) GFP-hVPS4(EQ) fluorescence. (C) Immunofluorescent stain against GFP. Arrows point to the GFP-hVPS4(EQ)-enriched TICS. (D) Image of YFP-TSG-3′ fluorescence.
FIG. 5.
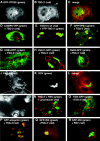
Colocalization of TSG-3′ with ESCRT and other cellular markers. (A-C) Cell transfected with a 1:1 ratio of GFP-VPS28 to TSG-3′. (A) Fluorescence image of GFP-VPS28 expression. (B) Immunofluorescence image of TSG-3′ expression. (C) Merge of panels A and B. (D, F-I, O-Q) Cells cotransfected with a 1:1 ratio of TSG-3′ and vectors expressing various fluorescently tagged cellular proteins. Each image is a merge of the TSG-3′ immunofluorescent stain image (red) and the fluorescent protein image (green). (D) CHMP6-YFP (ESCRT 3 protein). (F) GFP-hVPS4. (G) DsRed-CD63 (MVB marker). (H) Rab11-GFP (recycling endosome marker). (I) Rab9-GFP (late endosome marker). (O) TGN38-GFP (trans-Golgi marker). (P) GFP-ubiquitin (ubiquitin marker). (Q) GFP-250 (aggresome marker). (E) Cell transfected with a 1:1 ratio of YFP-TSG-3′ (green) to wild-type TSG101 (TSG101 wt) (red). (J-M) Cell transfected with a 1:1 ratio of RSV Gag-GFP to TSG-3′ treated with FM4-64 for the duration of the transfection (20 h). (J) Image of FM4-64 staining. (K) Image of RSV Gag-GFP expression (in TSG-3′ compartments). (L) Merge of panels J and K. (M) Blow up of the boxed location of panel J. (N) Cell transfected with a 1:1 ratio of RSV Gag-GFP (green) to TSG-3′ and stained live for 10 min with Lysotracker (red, acidic compartment marker). (R) Cell transfected with a 1:1 ratio of GFP-250 (green, aggresome marker) to RSV Gag-DsRed (red).
FIG. 6.
TEM and SEM analysis of TSG-3′ compartments. (A) Thin-section TEM of cell transfected with YFP-TSG-3′ and RSV Gag. (B-D) Cell transfected with a 1:1 ratio of YFP-TSG-3′ to RSV Gag. The cell was stripped of its membrane and actin cytoskeleton prior to imaging. (B) Fluorescence image of cellular remains and TSG-3′ compartments. (C-D) Low- and high-magnification SEM images of the same cellular remains.
Similar articles
- The functionally exchangeable L domains in RSV and HIV-1 Gag direct particle release through pathways linked by Tsg101.
Medina G, Zhang Y, Tang Y, Gottwein E, Vana ML, Bouamr F, Leis J, Carter CA. Medina G, et al. Traffic. 2005 Oct;6(10):880-94. doi: 10.1111/j.1600-0854.2005.00323.x. Traffic. 2005. PMID: 16138902 Free PMC article. - Defects in human immunodeficiency virus budding and endosomal sorting induced by TSG101 overexpression.
Goila-Gaur R, Demirov DG, Orenstein JM, Ono A, Freed EO. Goila-Gaur R, et al. J Virol. 2003 Jun;77(11):6507-19. doi: 10.1128/jvi.77.11.6507-6519.2003. J Virol. 2003. PMID: 12743307 Free PMC article. - Overexpression of the N-terminal domain of TSG101 inhibits HIV-1 budding by blocking late domain function.
Demirov DG, Ono A, Orenstein JM, Freed EO. Demirov DG, et al. Proc Natl Acad Sci U S A. 2002 Jan 22;99(2):955-60. doi: 10.1073/pnas.032511899. Proc Natl Acad Sci U S A. 2002. PMID: 11805336 Free PMC article. - [HIV budding and Tsg101].
Yasuda J. Yasuda J. Uirusu. 2005 Dec;55(2):281-6. doi: 10.2222/jsv.55.281. Uirusu. 2005. PMID: 16557014 Review. Japanese. - Novel Tsg101 Binding Partners Regulate Viral L Domain Trafficking.
Strickland M, Nyenhuis D, Watanabe SM, Tjandra N, Carter CA. Strickland M, et al. Viruses. 2021 Jun 15;13(6):1147. doi: 10.3390/v13061147. Viruses. 2021. PMID: 34203832 Free PMC article. Review.
Cited by
- FIV Gag: virus assembly and host-cell interactions.
Luttge BG, Freed EO. Luttge BG, et al. Vet Immunol Immunopathol. 2010 Mar 15;134(1-2):3-13. doi: 10.1016/j.vetimm.2009.10.003. Epub 2009 Oct 14. Vet Immunol Immunopathol. 2010. PMID: 19910057 Free PMC article. Review. - Distinct Roles of Cellular ESCRT-I and ESCRT-III Proteins in Efficient Entry and Egress of Budded Virions of Autographa californica Multiple Nucleopolyhedrovirus.
Yue Q, Yu Q, Yang Q, Xu Y, Guo Y, Blissard GW, Li Z. Yue Q, et al. J Virol. 2017 Dec 14;92(1):e01636-17. doi: 10.1128/JVI.01636-17. Print 2018 Jan 1. J Virol. 2017. PMID: 29046462 Free PMC article. - Recruitment of the oncoprotein v-ErbA to aggresomes.
Bondzi C, Brunner AM, Munyikwa MR, Connor CD, Simmons AN, Stephens SL, Belt PA, Roggero VR, Mavinakere MS, Hinton SD, Allison LA. Bondzi C, et al. Mol Cell Endocrinol. 2011 Jan 30;332(1-2):196-212. doi: 10.1016/j.mce.2010.10.012. Epub 2010 Nov 12. Mol Cell Endocrinol. 2011. PMID: 21075170 Free PMC article. - Tsg101 can replace Nedd4 function in ASV Gag release but not membrane targeting.
Medina G, Pincetic A, Ehrlich LS, Zhang Y, Tang Y, Leis J, Carter CA. Medina G, et al. Virology. 2008 Jul 20;377(1):30-8. doi: 10.1016/j.virol.2008.04.024. Virology. 2008. PMID: 18555885 Free PMC article. - The functionally exchangeable L domains in RSV and HIV-1 Gag direct particle release through pathways linked by Tsg101.
Medina G, Zhang Y, Tang Y, Gottwein E, Vana ML, Bouamr F, Leis J, Carter CA. Medina G, et al. Traffic. 2005 Oct;6(10):880-94. doi: 10.1111/j.1600-0854.2005.00323.x. Traffic. 2005. PMID: 16138902 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
- CA47482/CA/NCI NIH HHS/United States
- R01 CA047482/CA/NCI NIH HHS/United States
- R37 CA047482/CA/NCI NIH HHS/United States
- R01 CA020081/CA/NCI NIH HHS/United States
- CA20081/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Miscellaneous