Simple and highly efficient BAC recombineering using galK selection - PubMed (original) (raw)
Simple and highly efficient BAC recombineering using galK selection
Søren Warming et al. Nucleic Acids Res. 2005.
Abstract
Recombineering allows DNA cloned in Escherichia coli to be modified via lambda (lambda) Red-mediated homologous recombination, obviating the need for restriction enzymes and DNA ligases to modify DNA. Here, we describe the construction of three new recombineering strains (SW102, SW105 and SW106) that allow bacterial artificial chromosomes (BACs) to be modified using galK positive/negative selection. This two-step selection procedure allows DNA to be modified without introducing an unwanted selectable marker at the modification site. All three strains contain an otherwise complete galactose operon, except for a precise deletion of the galK gene, and a defective temperature-sensitive lambda prophage that makes recombineering possible. SW105 and SW106 cells in addition carry l-arabinose-inducible Cre or Flp genes, respectively. The galK function can be selected both for and against. This feature greatly reduces the background seen in other negative-selection schemes, and galK selection is considerably more efficient than other related selection methods published. We also show how galK selection can be used to rapidly introduce point mutations, deletions and loxP sites into BAC DNA and thus facilitate functional studies of SNP and/or disease-causing point mutations, the identification of long-range regulatory elements and the construction of conditional targeting vectors.
Figures
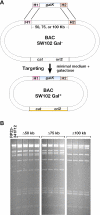
Figure 1
Sequence analysis of the galactose operon in strains DY380 (A), SW101 (B) and SW102 (C). In SW102, the ORF of galK was deleted, leaving only 33 bp of galK behind to make sure that translation of galM is initiated properly. EcoRI: the restriction site used to clone the 5′ and 3′ homology arms flanking galK.
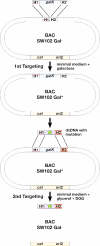
Figure 2
Overview of the galK selection scheme. The result of the first targeting event is the insertion of constitutively active galK into a defined position on the BAC by selection on minimal medium containing galactose and chloramphenicol to select for the maintenance of the BAC. The bacteria are now phenotypically Gal+. Next, the galK cassette is replaced by a dsDNA oligo, a PCR product, or a cloned dsDNA fragment carrying a desired mutation (indicated by a star) and flanked by the same homology arms used in the first selection step. This is achieved by negative selection using minimal medium containing 2-deoxy-galactose (DOG) with glycerol as the sole carbon source. The bacteria become phenotypically Gal−. H1 and H2, homology arms 1 and 2, respectively; cat, chloramphenicol acetyl transferase gene; ori2, BAC origin of replication; galK, E.coli galactokinase gene driven by a minimal promoter.
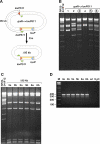
Figure 3
Introduction of a G12D mutation in the Nras gene. (A) SpeI restriction analysis of BAC miniprep DNA. First lane is the unmodified CITB-50J2 Nras BAC. Lanes 1–12 show digestion patterns of 12 clones counterselected for the substitution of galK with an oligo containing the G→A substitution for the second position of codon 12 of Nras. Clones 7 and 10 had internal deletions, indicating that DOG resistance was achieved by spontaneous deletion and not homologous recombination. These two clones were not analyzed further. (B) Sequence analysis of a PCR product spanning the modified region from clones 1–6, 8–9 and 11–12. All clones had the intended substitution (highlighted). However, clones 9 and 11 also had an internal basepair deletion indicated by a minus (highlighted). The Nras ATG and codon 12 are indicated (shadow).
Figure 4
BAC trimming using galK selection. (A) Illustration of the design of the deletion experiment. Homology arm 1 (H1) was held constant, and H2 was separated from H1 by either 50, 75 or 100 kb. (B) SpeI restriction analysis of BAC miniprep DNA from 12 clones showing deletions of 50, 75 and 100 kb, respectively, after the insertion of the galK selection cassette. The first lane is unmodified RP23-341F12 BAC DNA, which was included as a control. All tested clones had the intended deletion.
Figure 5
Insertion of a loxP511 site. (A) The location of the wild-type and mutant loxP sites in the BAC backbone are indicated along with the extra mutant loxP511 site that was introduced into the BAC genomic insert via galK counterselection. The 95 kb region deleted by Cre-mediated recombination between the two loxP511 sites is indicated, and PCR primers used to confirm the deletion are shown as small arrows. (B) SpeI restriction analysis of six miniprep clones selected for the replacement of galK with a dsDNA oligo containing the mutant loxP511 site. Clones 3, 5 and 6 (circles) had the same restriction pattern as the unmodified BAC, indicating that DOG resistance occurred due to the intended homologous recombination event. Clones 1, 2 and 4 had large deletions and were not analyzed further. (C) SpeI restriction analysis of BAC miniprep DNA from clones 3, 5 and 6 after transformation into Cre-induced EL350 cells. Two clones from each parental clone were tested. The restriction pattern shows that the 95 kb region flanked by two loxP511 sites is deleted from all clones analyzed, confirming the correct insertion of loxP511 in clones 3, 5 and 6. (D) PCR analysis of the six clones from (C) with one primer mapping to the BAC backbone and the other to a position distal to the inserted loxP511 site.
Figure 6
Same experiment as in Figure 5 with modifications as indicated at the top of each panel. (A) SpeI digest of 10 minipreps from a control experiment without heat-induction and without the loxP511 dsDNA oligo. (B) SpeI digest of 10 minipreps from a control experiment without heat-induction but with the loxP511 dsDNA oligo. (C) SpeI digest of 10 minipreps from a control experiment with heat-induction but without the loxP511 dsDNA oligo. (D) SpeI digest of 10 minipreps from an experiment with heat-induction and with the loxP511 dsDNA oligo (comparable with Figure 5B). Clones with the parental digestion pattern indicating DOG resistance due to homologous recombination (clones 33 and 35, circles) are only seen in (D). DOG resistance in all other clones likely occurred due to internal deletions of the BACs. (E) SpeI restriction analysis of BAC miniprep DNA from clones 33 and 35 after transformation into Cre-induced EL350 cells. Two clones from each parental clone were tested. The restriction pattern shows that the 95 kb region flanked by two loxP511 sites is deleted from all the clones analyzed, confirming the correct insertion of loxP511 in clones 33 and 35 (compare with Figure 5C).
Similar articles
- [Development of a new recombineering system by gap repair].
Li SH, Hong X, Yu M, Chen W, Huang CF, Zhou JG. Li SH, et al. Yi Chuan Xue Bao. 2005 May;32(5):533-7. Yi Chuan Xue Bao. 2005. PMID: 16018266 Chinese. - Development of a bacterial artificial chromosome (BAC) recombineering procedure using galK-untranslated region (UTR) for the mutation of diploid genes.
Dai G, Kim S, O'Callaghan DJ, Kim SK. Dai G, et al. J Virol Methods. 2012 Jun;182(1-2):18-26. doi: 10.1016/j.jviromet.2012.02.010. Epub 2012 Mar 8. J Virol Methods. 2012. PMID: 22407056 Free PMC article. - Efficient and seamless DNA recombineering using a thymidylate synthase A selection system in Escherichia coli.
Wong QN, Ng VC, Lin MC, Kung HF, Chan D, Huang JD. Wong QN, et al. Nucleic Acids Res. 2005 Mar 30;33(6):e59. doi: 10.1093/nar/gni059. Nucleic Acids Res. 2005. PMID: 15800210 Free PMC article. - [Recombineering and its application].
Zhou JG, Hong X, Huang CF. Zhou JG, et al. Yi Chuan Xue Bao. 2003 Oct;30(10):983-8. Yi Chuan Xue Bao. 2003. PMID: 14669518 Review. Chinese. - [Red/ET recombination and its biomedical applications].
Wang JP, Zhang YM. Wang JP, et al. Sheng Wu Gong Cheng Xue Bao. 2005 May;21(3):502-6. Sheng Wu Gong Cheng Xue Bao. 2005. PMID: 16108384 Review. Chinese.
Cited by
- Efficient Bacterial Genome Engineering throughout the Central Dogma Using the Dual-Selection Marker tetA OPT.
Bayer CN, Sepulchro AGV, Rennig M, Nørholm MHH. Bayer CN, et al. ACS Synth Biol. 2022 Oct 21;11(10):3440-3450. doi: 10.1021/acssynbio.2c00345. Epub 2022 Oct 7. ACS Synth Biol. 2022. PMID: 36206506 Free PMC article. - An essential role for γ-herpesvirus latency-associated nuclear antigen homolog in an acute lymphoproliferative disease of cattle.
Palmeira L, Sorel O, Van Campe W, Boudry C, Roels S, Myster F, Reschner A, Coulie PG, Kerkhofs P, Vanderplasschen A, Dewals BG. Palmeira L, et al. Proc Natl Acad Sci U S A. 2013 May 21;110(21):E1933-42. doi: 10.1073/pnas.1216531110. Epub 2013 Apr 29. Proc Natl Acad Sci U S A. 2013. PMID: 23630278 Free PMC article. - A temporal gate for viral enhancers to co-opt Toll-like-receptor transcriptional activation pathways upon acute infection.
Kropp KA, Hsieh WY, Isern E, Forster T, Krause E, Brune W, Angulo A, Ghazal P. Kropp KA, et al. PLoS Pathog. 2015 Apr 9;11(4):e1004737. doi: 10.1371/journal.ppat.1004737. eCollection 2015 Apr. PLoS Pathog. 2015. PMID: 25856589 Free PMC article. - The GABRG2 nonsense mutation, Q40X, associated with Dravet syndrome activated NMD and generated a truncated subunit that was partially rescued by aminoglycoside-induced stop codon read-through.
Huang X, Tian M, Hernandez CC, Hu N, Macdonald RL. Huang X, et al. Neurobiol Dis. 2012 Oct;48(1):115-23. doi: 10.1016/j.nbd.2012.06.013. Epub 2012 Jun 30. Neurobiol Dis. 2012. PMID: 22750526 Free PMC article. - Physiology of Highly Radioresistant Escherichia coli After Experimental Evolution for 100 Cycles of Selection.
Bruckbauer ST, Martin J, Minkoff BB, Veling MT, Lancaster I, Liu J, Trimarco JD, Bushnell B, Lipzen A, Wood EA, Sussman MR, Pennacchio C, Cox MM. Bruckbauer ST, et al. Front Microbiol. 2020 Sep 22;11:582590. doi: 10.3389/fmicb.2020.582590. eCollection 2020. Front Microbiol. 2020. PMID: 33072055 Free PMC article.
References
- Yang X.W., Model P., Heintz N. Homologous recombination based modification in Escherichia coli and germline transmission in transgenic mice of a bacterial artificial chromosome. Nat. Biotechnol. 1997;15:859–865. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous