N-myristoylation determines dual targeting of mammalian NADH-cytochrome b5 reductase to ER and mitochondrial outer membranes by a mechanism of kinetic partitioning - PubMed (original) (raw)
N-myristoylation determines dual targeting of mammalian NADH-cytochrome b5 reductase to ER and mitochondrial outer membranes by a mechanism of kinetic partitioning
Sara Colombo et al. J Cell Biol. 2005.
Abstract
Mammalian NADH-cytochrome b5 reductase (b5R) is an N-myristoylated protein that is dually targeted to ER and mitochondrial outer membranes. The N-linked myristate is not required for anchorage to membranes because a stretch of hydrophobic amino acids close to the NH2 terminus guarantees a tight interaction of the protein with the phospholipid bilayer. Instead, the fatty acid is required for targeting of b5R to mitochondria because a nonmyristoylated mutant is exclusively localized to the ER. Here, we have investigated the mechanism by which N-linked myristate affects b5R targeting. We find that myristoylation interferes with interaction of the nascent chain with signal recognition particle, so that a portion of the nascent chains escapes from cotranslational integration into the ER and can be post-translationally targeted to the mitochondrial outer membrane. Thus, competition between two cotranslational events, binding of signal recognition particle and modification by N-myristoylation, determines the site of translation and the localization of b5R.
Figures
Figure 1.
Schematic representation of the NH 2 -terminal regions of the three constructs used in this study. (A) The residues that constitute the myristoylation consensus are in boldface, the Gly that accepts the myristoyl moiety is boxed. This residue is mutated to Ala in G2A b5R. The rectangle downstream to Arg10 represents the hydrophobic region in all three constructs; this region is lengthened by five residues (filled part of the rectangle) in b5Rext. (B) Amino acid sequence (one-letter code) of the hydrophobic region (flanked by a basic residue on both sides) of the three constructs. The residues inserted in b5Rext are shown in boldface. The average hydrophobicity of the 12 residues after Arg10 was calculated according to the STA PRIFT scale (Cornette et al., 1987), and is displayed in italics to the right of the sequences.
Figure 2.
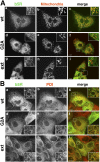
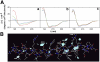
Dual localization of wt b5R, and ER-restricted distribution of G2A and b5Rext mutants, in stably transfected MDCK cells. Cells expressing each of the three proteins, as indicated to the left of the panels, were doubly stained with goat anti-b5R antibodies, followed by biotinylated anti–goat IgG and AlexaFluor 488–conjugated streptavidin (a, d, and g in A and B), and polyclonal anti-complex III antibodies or anti-protein disulfide isomerase (PDI) antibodies (b, e, and h in A and B respectively), both followed by Texas red–conjugated anti–rabbit IgG. In the merged images (shown in c, f, and i of both panels), the yellow color indicates colocalization. wt b5R colocalizes with the mitochondrial marker, but in addition shows a more widespread distribution (A), due to its ER localization demonstrated by colocalization with PDI (B). G2A and b5Rext do not show colocalization with the mitochondrial marker (A), but extensively colocalize with PDI (B). Bars, 10 μm. The closed boxes in the merged images indicate the area that is enlarged in the corresponding inset.
Figure 3.
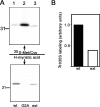
Analysis of myristoylation of b5R forms by metabolic labeling. MDCK cells expressing the indicated b5R forms were incubated with either 0.1 mCi/ml of 35S-Promix for 3 h (top) or 0.1 mCi/ml of 3H-myristic acid for 6 h. b5R was immunoprecipitated from the detergent lysates and analyzed by 11% SDS-PAGE fluorography. Both wt b5R (lane 1) and its extended mutant (lane 3) are myristoylated, whereas G2A (lane 2) is not. The position of the 31-kD size marker is shown. (B) b5Rext is less efficiently myristoylated than the wt protein. The band intensities of the gels of B were quantified by scanning. The 3H/35S ratio for wt b5R was arbitrarily set to 1.
Figure 4.
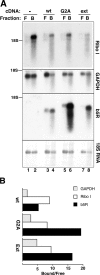
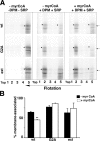
b5R mRNA is distributed on both free and bound polysomes, whereas mutant transcripts are translated exclusively on bound polysomes. (A) Blot analysis of RNA extracted from free (F) and bound (B) polysome fractions prepared from nontransfected MDCK (lanes 1 and 2) or from cells stably transfected with wt b5R (lanes 3 and 4), G2A (lanes 5 and 6), and b5Rext (lanes 7 and 8). 3.6 μg RNA from free and 1.8 μg from the bound polysomes were loaded. The blot was stained with methylene blue and then sequentially hybridized with probes for ribophorin I, GAPDH, and b5R as indicated. The bottom panel shows the methylene blue–stained 18S ribosomal RNA. The position of the 18S RNA in all panels is indicated. (B) Quantification by phosphorimaging of the signals of the experiment illustrated in A. The ratios of the intensities of the signal in the bound versus the free polysome fraction are shown after correction for the different RNA loads.
Figure 5.
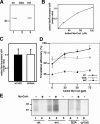
b5R and its mutant forms interact with SRP. (A) Synthetic transcripts coding for the indicated b5R forms were translated in wheat germ extract for 20 min in the absence or presence of 100 nM SRP, and analyzed by 11% SDS-PAGE autoradiography. SRP inhibits translation of the wt, G2A, and extended b5R (lanes 1–6), while having much less effect on the translation of a soluble b5R form that lacks the hydrophobic anchor (sol, lanes 7 and 8). (B) Dependence of translational slow-down on SRP concentration. wt b5R (top), G2A (middle), and b5Rext (bottom) transcripts were translated together with luciferase mRNA for 40 min in the presence of the indicated concentrations of SRP, and were analyzed by 11% SDS-PAGE phosphorimaging. The arrow and arrowhead indicate b5R and luciferase, respectively. (C) The intensities of the bands in six different experiments like the one shown in B were quantified. For each b5R form and in each experiment, the ratio of intensities of b5R to luciferase in the absence of SRP was set to 100% translation efficiency. Shown are averages with SEM.
Figure 6.
Myristoylation of b5R forms in the wheat germ extract and effect of myristoylation on interaction with SRP. (A) wt b5R (lane 1), G2A (lane 2), and b5Rext (lane 3) were translated in the presence of unlabeled Met and 3H-MyrCoA (see Materials and methods for details). Immunoprecipitates were run on 11% SDS–polyacrylamide gels, followed by blotting and phosphorimaging analysis. wt b5R and its extended mutant are myristoylated, whereas G2A is not. (B) Stoichiometry of in vitro myristoylation of wt b5R determined by double labeling. See Materials and methods for details on the experimental procedure. Calculation of the molar ratio of myristate to protein is based on the known specific radioactivities of the added compounds, and on the number of Met residues in the b5R sequence (8, not considering Met1). Any dilution of the specific radioactivity of 3H-MyrCoA or 35S-Met by the endogenous compounds is not considered. (C) Comparison of stoichiometry of in vitro myristoylation of wt b5R and b5Rext shows equal efficiency for the two proteins. wt b5R and b5Rext transcripts were translated in the wheat germ extract in the presence of 35S-Met and 3H-MyrCoA (21 μM). Calculation of the molar ratio of myristate to translated protein was as in B. Bars indicate the SEM (n = 5). (D) MyrCoA blunts the effect of SRP on the translation of wt b5R, but not of G2A and b5Rext. Transcripts coding for each of the three b5R forms were translated in wheat germ extract together with luciferase mRNA, 50 nM SRP, 35S-Met, and the indicated concentrations of unlabeled MyrCoA. For each concentration of MyrCoA, translation efficiency is the ratio of b5R band intensity to that of luciferase in the presence of SRP normalized to the same ratio in the absence of SRP, which was set at 100. Bars indicate the SEM (n = 5). (E) Inhibition by MyrCoA of the association of wt b5R nascent chains with SRP, assessed by cross-linking. Truncated mRNAs coding for the first 108 amino acids of wt b5R (lanes 1 and 2) and G2A (lanes 6 and 7), the first 113 amino acids of b5Rext (lanes 3–5), and the first 125 residues of cytochrome b(5) (lane 8) were translated for 30 min in reticulocyte lysate with or without 75 μM MyrCoA, as indicated. RNCs were recovered by centrifugation through a high salt sucrose cushion as detailed in the Materials and methods section. For each construct, equal amounts of TCA precipitable radioactivity were cross-linked with DSS and then immunoprecipitated with an anti-SRP54 antibody, with the exception of lane 5, in which precipitation was performed with a nonimmune serum. Adducts corresponding to cross-linked products of SRP and the nascent polypeptide chains of the three forms of reductase, but not of cytochrome b(5), were detected when anti-SRP54 was used for the immunoprecipitation. Note the strong inhibitory effect of added MyrCoA on wt b5R nascent chain cross-linking (lane 2), but not on the mutant b5R forms. Exposure times were 3 d for lanes 1–5 and 12 d for lanes 6–8.
Figure 7.
Myristoylation inhibits recruitment of b5R-synthesizing polysomes to ER membranes. (A) The truncated mRNAs coding for the NH2-terminal portion of the three b5R forms (described in the legend to Fig. 6 E) were translated in wheat germ extract without other additions (left column), or with the addition of DPM + SRP and MyrCoA, as indicated (see Materials and methods). The translated samples were brought to 1.8 M sucrose and run on high salt-sucrose flotation gradients. TCA-precipitated fractions from the gradients were analyzed by 14% SDS-PAGE phosphorimaging. Fraction 2 contains the 0.3/1.6 M sucrose interface. Lane 5 contains the bottom fraction plus the pellet. Note the shift of the nascent chains from the bottom of the gradient, containing the free polysomes, to the 0.3/1.6 M interface, when DPM were added. When MyrCoA was present, wt b5R nascent chains were partially shifted back to the free polysome fraction. The arrow on the right indicates the position of the 14-kD size marker. (B) Quantification of three independent experiments like the one of A. The bands indicated by the asterisk in lanes 2 and 5 were quantified. Shown is the percentage recovered in lane 2 with respect to the sum of the intensities in lanes 2 + 5. Bars indicate the SEM. **, highly significant difference between the − and + MyrCoA samples (P = 0.0061 by t test).
Figure 8.
Myristate stabilizes the α-helical conformation of b5R's NH2-terminal peptide. (A) CD spectra of b5R NH2-terminal peptide (G2-A33). Spectra were taken for the unmodified (blue), the N-myristoylated (red), or the N-acetylated (green) peptide, diluted in water (a), 50% TFE (b), or 25 mM SDS (c). In aqueous solution the N-myristoylated peptide has a spectrum indicative of a higher α-helical content than the other two peptides. (B) Result of GRID analysis with the methyl group probe. The b5R G2-A33 peptide in α-helical conformation is represented in wireframe. Contour surfaces with favorable interaction energies of the methyl probe (threshold = −10 kJ/mol) are shown in light blue.
Figure 9.
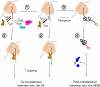
A kinetic partitioning model to explain the dual ER/MOM targeting of b5R. The different steps of alternative pathways are numbered and explained in the text. SRP is represented by the brown elongated body. The green segment of the nascent chain represents the myristoylation consensus, the red segment the hydrophobic, SRP-interacting region. The wavy black line represents the myristoyl moiety attached to the NH2 terminus of the nascent chain. The two blue forms in pathway 4 depict unknown chaperones that might be involved in the post-translational targeting of b5R to the MOM. See Discussion for further explanation.
Similar articles
- A role for N-myristoylation in protein targeting: NADH-cytochrome b5 reductase requires myristic acid for association with outer mitochondrial but not ER membranes.
Borgese N, Aggujaro D, Carrera P, Pietrini G, Bassetti M. Borgese N, et al. J Cell Biol. 1996 Dec;135(6 Pt 1):1501-13. doi: 10.1083/jcb.135.6.1501. J Cell Biol. 1996. PMID: 8978818 Free PMC article. - NADH-cytochrome b5 reductase and cytochrome b5 isoforms as models for the study of post-translational targeting to the endoplasmic reticulum.
Borgese N, D'Arrigo A, De Silvestris M, Pietrini G. Borgese N, et al. FEBS Lett. 1993 Jun 28;325(1-2):70-5. doi: 10.1016/0014-5793(93)81416-w. FEBS Lett. 1993. PMID: 8513896 Review. - Dual subcellular distribution of cytochrome b5 in plant, cauliflower, cells.
Zhao J, Onduka T, Kinoshita JY, Honsho M, Kinoshita T, Shimazaki K, Ito A. Zhao J, et al. J Biochem. 2003 Jan;133(1):115-21. doi: 10.1093/jb/mvg009. J Biochem. 2003. PMID: 12761206 - The carboxy-terminal 10 amino acid residues of cytochrome b5 are necessary for its targeting to the endoplasmic reticulum.
Mitoma J, Ito A. Mitoma J, et al. EMBO J. 1992 Nov;11(11):4197-203. doi: 10.1002/j.1460-2075.1992.tb05513.x. EMBO J. 1992. PMID: 1396600 Free PMC article. - Regulation of mammalian desaturases by myristic acid: N-terminal myristoylation and other modulations.
Rioux V, Pédrono F, Legrand P. Rioux V, et al. Biochim Biophys Acta. 2011 Jan;1811(1):1-8. doi: 10.1016/j.bbalip.2010.09.005. Epub 2010 Oct 1. Biochim Biophys Acta. 2011. PMID: 20920594 Review.
Cited by
- Privileged proteins with a second residence: dual targeting and conditional re-routing of mitochondrial proteins.
Pines O, Horwitz M, Herrmann JM. Pines O, et al. FEBS J. 2024 Dec;291(24):5379-5393. doi: 10.1111/febs.17191. Epub 2024 Jun 10. FEBS J. 2024. PMID: 38857249 Free PMC article. Review. - Protein lipidation in the tumor microenvironment: enzymology, signaling pathways, and therapeutics.
Xu M, Xu B. Xu M, et al. Mol Cancer. 2025 May 7;24(1):138. doi: 10.1186/s12943-025-02309-7. Mol Cancer. 2025. PMID: 40335986 Free PMC article. Review. - ER-SURF: Riding the Endoplasmic Reticulum Surface to Mitochondria.
Koch C, Schuldiner M, Herrmann JM. Koch C, et al. Int J Mol Sci. 2021 Sep 6;22(17):9655. doi: 10.3390/ijms22179655. Int J Mol Sci. 2021. PMID: 34502567 Free PMC article. Review. - Mouse Stbd1 is _N_-myristoylated and affects ER-mitochondria association and mitochondrial morphology.
Demetriadou A, Morales-Sanfrutos J, Nearchou M, Baba O, Kyriacou K, Tate EW, Drousiotou A, Petrou PP. Demetriadou A, et al. J Cell Sci. 2017 Mar 1;130(5):903-915. doi: 10.1242/jcs.195263. Epub 2017 Jan 30. J Cell Sci. 2017. PMID: 28137759 Free PMC article.
References
- Adams, J.M., and S. Cory. 2001. Life-or-death decisions by the Bcl-2 protein family. Trends Biochem. Sci. 26:61–66. - PubMed
- Bernardi, P., and G.F. Azzone. 1981. Cytochrome c as an electron shuttle between the outer and inner mitochondrial membranes. J. Biol. Chem. 256:7187–7192. - PubMed
- Borgese, N., and S. Gaetani. 1980. Site of synthesis of rat liver NADH-cytochrome b5 reductase, an integral membrane protein. FEBS Lett. 112:216–220. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources