Sequestration of TRAF2 into stress granules interrupts tumor necrosis factor signaling under stress conditions - PubMed (original) (raw)
Sequestration of TRAF2 into stress granules interrupts tumor necrosis factor signaling under stress conditions
Woo Jae Kim et al. Mol Cell Biol. 2005 Mar.
Abstract
The cellular stress response (SR) is a phylogenetically conserved protection mechanism that involves inhibition of protein synthesis through recruitment of translation factors such as eIF4G into insoluble stress granules (SGs) and blockade of proinflammatory responses by interruption of the signaling pathway from tumor necrosis factor alpha (TNF-alpha) to nuclear factor-kappaB (NF-kappaB) activation. However, the link between these two physiological phenomena has not been clearly elucidated. Here we report that eIF4GI, which is a scaffold protein interacting with many translation factors, interacts with TRAF2, a signaling molecule that plays a key role in activation of NF-kappaB through TNF-alpha. These two proteins colocalize in SGs during cellular exposure to stress conditions. Moreover, TRAF2 is absent from TNFR1 complexes under stress conditions even after TNF-alpha treatment. This suggests that stressed cells lower their biological activities by sequestration of translation factors and TRAF2 into SGs through a protein-protein interaction.
Figures
FIG. 1.
eIF4GI interacts with TRAF2. (A) 293T cells were transfected with plasmids expressing GST (lanes 1 and 3), GST-tagged eIF4GIΔC (lanes 2 and 4), and the HA-tagged TRAF and Finger domains of TRAF2 (panels labeled “HA-Finger domain” and “HA-TRAF domain”). The input panels show proteins in the cell extract before immunoprecipitation (1/50 of lysatens used in immunoprecipitation). Lanes 3 and 4 show proteins precipitated by GST-resin. WB, Western blotting. (B) HeLa cell extracts were subjected to immunoprecipitation using a rabbit preimmune serum (lane 2) or anti-eIF4GI antibody (lane3) and then analyzed by WB with antibodies against TRAF2 (panel labeled “TRAF2”), PABP (panel labeled “PABP”), and eIF4GI (panel labeled “eIF4GI”). (C) 293T cells were transfected with plasmids expressing FLAG or FLAG-tagged TRAF1, TRAF2, TRAF3, TRAF5, or TRAF6. Cell lysates were subjected to immunoprecipitation using an anti-FLAG monoclonal antibody and then analyzed by WB using anti-eIF4GI or eIF4GII antibodies. (D) Y2H analysis of TRAF2-binding region in eIF4GI. The boxed numbers indicate the amino acid number in eIF4GI included in the bait vector. Positive and negative results in the Y2H analyses are indicated by “Yes” and “No,” respectively. (E) TRAF2-binding motifs in CD30, TNFRII, and eIF4GI. (F) 293T cells were transfected with plasmids expressing GST (lanes 1 and 5), GST-tagged eIF4GIΔC (lanes 2 and 6), GST-eIF4GI (293-303) (lanes 3 and 7), and mutant GST-eIF4GI (293-303) containing an AVEA sequence instead of SVEE in the consensus motif (lanes 4 and 8). Lanes 1 to 4 and 5 to 8 show proteins in the cell extract before and after precipitation with GST-resin, respectively.
FIG. 2.
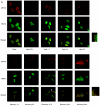
TRAF2 and eIF4GI migrate to SGs under heat stress conditions. (A) HeLa cells were heat treated at 44°C and then allowed to recover at 37°C for the indicated times. After treatments, cells were fixed, permeabilized, and treated with primary antibodies (an anti-eIF4GI polyclonal antibody and an anti-TRAF2 monoclonal antibody) and secondary antibodies (a rhodamine-conjugated donkey anti-rabbit immunoglobulin G and a fluorescein isothiocyanate-conjugated donkey anti-mouse immunoglobulin G). The localization patterns of eIF4GI and TRAF2 are shown in red and green, respectively. The boxes on the right side of the panels labeled “Heat 3 h” and “Recovery 48 h” show enlarged parts of pictures indicated by white boxes. (B) The same sets of samples shown in panel A were visualized with an anti-TIA-1 and an anti-eIF4GI antibody. (C) Distribution patterns of lamin A/C and eIF4GI before (Mock) and after (Heat 3 h) heat treatment were visualized with an anti-lamin A/C and an anti-eIF4GI antibody.
FIG. 2.
TRAF2 and eIF4GI migrate to SGs under heat stress conditions. (A) HeLa cells were heat treated at 44°C and then allowed to recover at 37°C for the indicated times. After treatments, cells were fixed, permeabilized, and treated with primary antibodies (an anti-eIF4GI polyclonal antibody and an anti-TRAF2 monoclonal antibody) and secondary antibodies (a rhodamine-conjugated donkey anti-rabbit immunoglobulin G and a fluorescein isothiocyanate-conjugated donkey anti-mouse immunoglobulin G). The localization patterns of eIF4GI and TRAF2 are shown in red and green, respectively. The boxes on the right side of the panels labeled “Heat 3 h” and “Recovery 48 h” show enlarged parts of pictures indicated by white boxes. (B) The same sets of samples shown in panel A were visualized with an anti-TIA-1 and an anti-eIF4GI antibody. (C) Distribution patterns of lamin A/C and eIF4GI before (Mock) and after (Heat 3 h) heat treatment were visualized with an anti-lamin A/C and an anti-eIF4GI antibody.
FIG. 3.
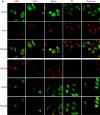
Effects of heat shock inducers on localization of eIF4GI and TRAF2. (A) HeLa cells grown on coverslips were treated with cadmium chloride (500 μM, 30 min), hydrogen peroxide (1 mM, 30 min), magnesium chloride (400 μM, 30 min), sodium arsenite (400 μM, 30 min), and puromycin (20 mg/ml, 6 h). Green fluorescence indicates endogenous TRAF2 (top panel) or eIF4GI (bottom panel), and red fluorescence indicates endogenous eIF4GI (top panel) or TIA-1 (bottom panel), respectively. Endogenous proteins were visualized as described in the legend to Fig. 2. (B) HeLa cells were heat treated as described in the legend to Fig. 2A and then analyzed by WB with antibodies against phospho-eIF2α and actin. (C) HeLa cells were treated with various stress-inducing agents and then analyzed by WB with antibodies against phospho-eIF2α and actin. (D) HeLa cells were mock treated (top panels) or treated with 10 μM of FCCP for 90 min (bottom panels) and then subjected to immunocytochemistry using anti-TRAF2 and -eIF4GI antibodies. TRAF2 and eIF4GI are shown in green and red, respectively. The nucleus is shown in blue by DAPI (4′,6′-diamidino-2-phenylindole) staining. (E) Solubility of proteins under normal and stress conditions. HeLa cells were mock treated (lanes 1 and 3) or heat treated for 3 h (lanes 2 and 4). Fractionation of the HeLa cell extract was performed as described elsewhere (10) with the addition of pretreatment with cross-linking agent 1,5-difluoro-2,4-dinitrobenzene (Sigma). The soluble (lanes 1 and 2) and insoluble (lanes 3 and 4) fractions were analyzed by Western blotting with antibodies against eIF4GI (panel labeled “eIF4GI”), PABP (panel labeled “PABP”), and TRAF2 (panel labeled “TRAF2”).
FIG. 3.
Effects of heat shock inducers on localization of eIF4GI and TRAF2. (A) HeLa cells grown on coverslips were treated with cadmium chloride (500 μM, 30 min), hydrogen peroxide (1 mM, 30 min), magnesium chloride (400 μM, 30 min), sodium arsenite (400 μM, 30 min), and puromycin (20 mg/ml, 6 h). Green fluorescence indicates endogenous TRAF2 (top panel) or eIF4GI (bottom panel), and red fluorescence indicates endogenous eIF4GI (top panel) or TIA-1 (bottom panel), respectively. Endogenous proteins were visualized as described in the legend to Fig. 2. (B) HeLa cells were heat treated as described in the legend to Fig. 2A and then analyzed by WB with antibodies against phospho-eIF2α and actin. (C) HeLa cells were treated with various stress-inducing agents and then analyzed by WB with antibodies against phospho-eIF2α and actin. (D) HeLa cells were mock treated (top panels) or treated with 10 μM of FCCP for 90 min (bottom panels) and then subjected to immunocytochemistry using anti-TRAF2 and -eIF4GI antibodies. TRAF2 and eIF4GI are shown in green and red, respectively. The nucleus is shown in blue by DAPI (4′,6′-diamidino-2-phenylindole) staining. (E) Solubility of proteins under normal and stress conditions. HeLa cells were mock treated (lanes 1 and 3) or heat treated for 3 h (lanes 2 and 4). Fractionation of the HeLa cell extract was performed as described elsewhere (10) with the addition of pretreatment with cross-linking agent 1,5-difluoro-2,4-dinitrobenzene (Sigma). The soluble (lanes 1 and 2) and insoluble (lanes 3 and 4) fractions were analyzed by Western blotting with antibodies against eIF4GI (panel labeled “eIF4GI”), PABP (panel labeled “PABP”), and TRAF2 (panel labeled “TRAF2”).
FIG. 4.
TRAF2 forms different complexes under normal and stress conditions. (A) HeLa cells were mock treated (lanes 1 and 2) or heat treated for 3 h (lanes 3 and 4) and then treated with 100 ng of human TNF-α/ml for 1 h (lanes 2 and 4). The samples in lanes 1 and 3 were not treated with TNF-α. After TNF-α treatment, cells were washed with cold PBS and then subjected to immunoprecipitation with a monoclonal anti-TNFR1 antibody. Control proteins (actin and lamin A/C) are shown in panels actin and lamin A/C. (B) HeLa cells were mock treated (top panels) or heat-treated for 1 h (bottom panels) and then subjected to immunocytochemistry using anti-RIP and anti-TRAF2 antibodies. TRAF2 and RIP are shown in green and red, respectively. The nucleus is shown in blue by DAPI staining.
FIG. 5.
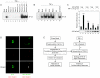
eIF4GI-TRAF2 interaction blocks TNF signaling. (A) HeLa cells were treated as described in the legend to Fig. 2A and then further treated with 100 ng of TNF-α/ml at 37°C for 1 h. EMSA was performed using 32P-labeled oligonucleotides corresponding to the NF-κB-responsive element of the c-IAP2 promoter (17). (B) HeLa cells were treated as described in the legend to Fig. 3A and then further treated with 100 ng of TNF-α/ml at 37°C for 1 h. EMSA was performed as described for panel A. (C) 293T cells were cotransfected with pNF-κB-Luc (Stratagene), which encodes the firefly luciferase gene under the control of a NF-κB response element, and various amounts of plasmid pEBG-eIF4GI, which produces GST-eIF4GI. Total amounts of effector plasmids were kept the same (9 μg) in all experiments by adjusting the amounts of control plasmid pEBG that was cotransfected with plasmid pEBG-eIF4GI. Control plasmid pRLCMV-Luc (Promega) was also cotransfected to control for transfection efficiency. After 48 h, cells were incubated in serum-free medium for 3 h and then further incubated for 12 h in the presence (lanes 2, 4, 6, and 8) and absence (lanes 1, 3, 5, and 7) of 10 ng of TNF-α/ml. Three independent experiments were performed under each set of conditions. The bottom panel shows GST-eIF4GI polypeptide detected by Western blotting with an anti-GST antibody. (D) COS-7 cells were cotransfected with plasmids expressing GFP- and RFP-tagged TRAF2 (RFP-TRAF2) or GFP-tagged eIF4GI (GFP-eIF4GI) and RFP-TRAF2. (E) A hypothetical scheme for the modulation of translation and TNF signaling. The asterisk indicates that the interaction between TRAF2 and TNF-α receptor can be blocked by the sequestration of TRAF2 into SGs through an interaction with eIF4GI.
Similar articles
- Tumor necrosis factor (TNF) receptor-associated factor (TRAF)-interacting protein (TRIP) negatively regulates the TRAF2 ubiquitin-dependent pathway by suppressing the TRAF2-sphingosine 1-phosphate (S1P) interaction.
Park ES, Choi S, Shin B, Yu J, Yu J, Hwang JM, Yun H, Chung YH, Choi JS, Choi Y, Rho J. Park ES, et al. J Biol Chem. 2015 Apr 10;290(15):9660-73. doi: 10.1074/jbc.M114.609685. Epub 2015 Feb 25. J Biol Chem. 2015. PMID: 25716317 Free PMC article. - TRAF2 plays a key, nonredundant role in LIGHT-lymphotoxin beta receptor signaling.
Kim YS, Nedospasov SA, Liu ZG. Kim YS, et al. Mol Cell Biol. 2005 Mar;25(6):2130-7. doi: 10.1128/MCB.25.6.2130-2137.2005. Mol Cell Biol. 2005. PMID: 15743811 Free PMC article. - TRAF2 multitasking in TNF receptor-induced signaling to NF-κB, MAP kinases and cell death.
Borghi A, Verstrepen L, Beyaert R. Borghi A, et al. Biochem Pharmacol. 2016 Sep 15;116:1-10. doi: 10.1016/j.bcp.2016.03.009. Epub 2016 Mar 16. Biochem Pharmacol. 2016. PMID: 26993379 Review. - That which does not kill you makes you stronger: a molecular mechanism for preconditioning.
McDunn JE, Cobb JP. McDunn JE, et al. Sci STKE. 2005 Jul 5;2005(291):pe34. doi: 10.1126/stke.2912005pe34. Sci STKE. 2005. PMID: 15998871 Review.
Cited by
- Inactivation of the mTORC1-eukaryotic translation initiation factor 4E pathway alters stress granule formation.
Fournier MJ, Coudert L, Mellaoui S, Adjibade P, Gareau C, Côté MF, Sonenberg N, Gaudreault RC, Mazroui R. Fournier MJ, et al. Mol Cell Biol. 2013 Jun;33(11):2285-301. doi: 10.1128/MCB.01517-12. Epub 2013 Apr 1. Mol Cell Biol. 2013. PMID: 23547259 Free PMC article. - Regulated protein aggregation: stress granules and neurodegeneration.
Wolozin B. Wolozin B. Mol Neurodegener. 2012 Nov 20;7:56. doi: 10.1186/1750-1326-7-56. Mol Neurodegener. 2012. PMID: 23164372 Free PMC article. Review. - Stress Beyond Translation: Poxviruses and More.
Liem J, Liu J. Liem J, et al. Viruses. 2016 Jun 14;8(6):169. doi: 10.3390/v8060169. Viruses. 2016. PMID: 27314378 Free PMC article. Review. - Viral modulation of stress granules.
Valiente-Echeverría F, Melnychuk L, Mouland AJ. Valiente-Echeverría F, et al. Virus Res. 2012 Nov;169(2):430-7. doi: 10.1016/j.virusres.2012.06.004. Epub 2012 Jun 15. Virus Res. 2012. PMID: 22705970 Free PMC article. Review. - Chronic starvation induces noncanonical pro-death stress granules.
Reineke LC, Cheema SA, Dubrulle J, Neilson JR. Reineke LC, et al. J Cell Sci. 2018 Oct 5;131(19):jcs220244. doi: 10.1242/jcs.220244. J Cell Sci. 2018. PMID: 30185525 Free PMC article.
References
- Anderson, P., and N. Kedersha. 2002. Stressful initiations. J. Cell Sci. 115:3227-3234. - PubMed
- Arch, R. H., R. W. Gedrich, and C. B. Thompson. 2000. Translocation of TRAF proteins regulates apoptotic threshold of cells. Biochem. Biophys. Res. Commun. 272:936-945. - PubMed
- Back, S. H., J. E. Kim, J. Rho, B. Hahm, T. G. Lee, E. E. Kim, J. M. Cho, and S. K. Jang. 2000. Expression and purification of an active, full-length hepatitis C viral NS4A. Protein Expr. Purif. 20:196-206. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials