Atg17 functions in cooperation with Atg1 and Atg13 in yeast autophagy - PubMed (original) (raw)
Atg17 functions in cooperation with Atg1 and Atg13 in yeast autophagy
Yukiko Kabeya et al. Mol Biol Cell. 2005 May.
Abstract
In eukaryotic cells, nutrient starvation induces the bulk degradation of cellular materials; this process is called autophagy. In the yeast Saccharomyces cerevisiae, most of the ATG (autophagy) genes are involved in not only the process of degradative autophagy, but also a biosynthetic process, the cytoplasm to vacuole (Cvt) pathway. In contrast, the ATG17 gene is required specifically in autophagy. To better understand the function of Atg17, we have performed a biochemical characterization of the Atg17 protein. We found that the atg17delta mutant under starvation condition was largely impaired in autophagosome formation and only rarely contained small autophagosomes, whose size was less than one-half of normal autophagosomes in diameter. Two-hybrid analyses and coimmunoprecipitation experiments demonstrated that Atg17 physically associates with Atg1-Atg13 complex, and this binding was enhanced under starvation conditions. Atg17-Atg1 binding was not detected in atg13delta mutant cells, suggesting that Atg17 interacts with Atg1 through Atg13. A point mutant of Atg17, Atg17(C24R), showed reduced affinity for Atg13, resulting in impaired Atg1 kinase activity and significant defects in autophagy. Taken together, these results indicate that Atg17-Atg13 complex formation plays an important role in normal autophagosome formation via binding to and activating the Atg1 kinase.
Figures
Figure 1.
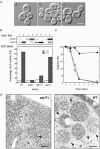
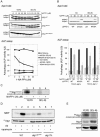
ATG17 is essential for normal autophagosome formation. (A) _ATG17_-null mutant appears devoid of autophagic bodies in the vacuole. Wild-type (KVY55; a) and _atg17_Δ (YYK382) strains transformed with either vector alone (b) or the multicopy ATG17 plasmid (c) were incubated for 4 h under nitrogen and carbon starvation condition in the presence of 1 mM PMSF. DIC images are shown. Bar, 5 μm. (B) The _atg17_Δ strain blocks autophagy. Wild-type (KVY55), _atg9_Δ (YTS007), _atg17_Δ (YYK382), and _atg11_Δ (YYK475) cells in YEPD (lanes 1, 3, 5, and 7) or in SD(-N) for 4 h (lanes 2, 4, 6, and 8) were analyzed by anti-Ape1 immunoblot (top). The autophagic activity was measured by the ALP assay before (□) and after (▪) nitrogen starvation for 6 h (bottom). (C) The _atg17_Δ strain is sensitive to nitrogen starvation. Wild-type (KA311A, •), _atg1_Δ (YYK36, ○), and _atg17_Δ (YYK111, □) cells were cultured in nitrogen starvation medium. Aliquots were removed at the indicated times and spread onto YEPD plates. The number of colonies was determined after 2–3 d. (D) Deletion of ATG17 causes formation of aberrantly small autophagosomes. The _pep4_Δ _atg17_Δ cells (YYK381; left) grown to log phase in YEPD were incubated in SD(-N) for 4 h and subjected to electron microscopy. Small autophagosome (opened arrowhead) and autophagic body (closed arrowhead) are detected in the cytoplasm and the vacuole (V), respectively. Note that the Cvt complex is contained in both autophagosome and autophagic body (left panel). Right panel shows normal autophagic bodies of wild-type cell. Bar, 200 nm.
Figure 2.
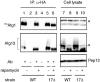
The interaction between Atg1 and Atg13 was not affected by atg17_Δ. HA_ATG1 integrated cells (wild-type [WT], YYK422; or _atg17_Δ [17Δ; YYK 416]) additionally expressing Atg13 via a low-copy plasmid were enzymatically converted to spheroplasts and treated with 0.2 μg/ml rapamycin for 1 h. The total lysates were immunoprecipitated with anti-HA antibody and immunoblotted with anti-HA and anti-Atg13 antibodies. The asterisks show nonspecific bands in the total lysate that were recognized by anti-HA and -Atg13 antibodies. Immunoblot with anti-Pep12 antibody (Sigma) was carried out to monitor amount of cell lysate.
Figure 3.
Atg17 interacts with Atg1 in an Atg13-dependent manner. Wild-type cells (YYK422), _atg1_Δ (YYK424) harboring a pATG13 CEN plasmid, and _atg13_Δ cells (YYK 426) were treated with rapamycin as shown in Figure 2. HAAtg1 was immunoprecipitated with anti-HA antibody and immunoblotted with anti-HA, Atg13, and Atg17 antibodies. Asterisks indicate nonspecific bands. Pep12 indicates that the amount of cell lysate per lane is equal.
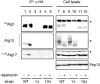
Figure 4.
Atg17 interacts with Atg13 under starvation conditions. Both wild-type (YYK422) and _atg1_Δ (YYK 424) cells, which harbor additionally expressed Atg13 via a low-copy plasmid, were integrated with FLAGAtg17. The cells were grown in YEPD medium and were treated with rapamycin. Atg13 was immunoprecipitated with anti-Atg13 antibody. Associated proteins were visualized by immunoblotting with anti-Atg13 and FLAG antibodies. The asterisks indicate nonspecific bands. Equal amount cell lysate was loaded on the gel by monitoring with Pep12.
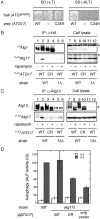
Figure 5.
A mutation in ATG17 abolishes complex formation and defects in autophagy. (A) atg17 mutant screened by the two-hybrid system. Plasmids transformed into PJ69–4A strains are indicated. The cells were grown on SC-Leu-Trp plate (left panel), replicaplated on SC-Ade-Leu-Trp plate (right panel), and then incubated at 30°C for 3 d. (B) Atg17C24R does not bind Atg1. Total lysates were prepared from wild-type cells (YYK422) and atg17C24R cells (YYK 467) chromosomally integrated with either HA_ATG1_ and FLAGATG17 (WT) or HA_ATG1_ and FLAGatg17C24R (CR). The cells were grown in YEPD in the absence or presence of rapamycin. The lysates were immunoprecipitated with anti-HA antibody and immunoblotted with anti-Atg17 and anti-HA antibodies. (C) Atg17C24R shows reduced affinity for Atg13. Immunoprecipitation with anti-Atg13 antibody was carried out as described in Figure 4 using wild-type cells (YYK422), atg17C24R cells (YYK 467) harboring pRS316[_ATG13_], or _atg13_Δ cells (YYK 426). After SDS-PAGE, immunoprecipitates were subjected to immunoblot analysis using anti-Atg13 or anti-FLAG antibody as indicated. The asterisk indicates a nonspecific band. (D) atg17C24R mutant is defective for autophagy. KVY55 cells (pho8::pho8_Δ_60) or YYK382 cells (_atg17_Δ pho8::pho8_Δ_60) harboring the wild-type ATG17 or atg17C24R mutant on 2-μ plasmids were grown to 1 OD600/ml in YEPD medium and then transferred to SD(-N) medium. Lysates from the cells before (0 h, □) and after 6-h starvation (▪) were used for the ALP assay. The error bars indicate the SD of three independent experiments.
Figure 6.
Interaction between Atg13 and Atg17 is required for Atg1 kinase activity. (A) Chemical genetic analysis of ATG1 using YEPD-grown cells. Cells (YYK126, _atg1_Δ _pho8_Δ60) harboring the indicated ATG1 plasmids were grown in YEPD medium, and the cells were pretreated with 1-NA-PP1 (DMSO solution) at the indicated concentrations for 1 h. The control cells were treated with DMSO. The cells were then transferred to SD(-N) medium containing the same concentration of 1-NA-PP1 or they continued growing in YEPD for 4 h. The cells were collected and subjected to Ape1 blotting (top) or ALP assay (bottom). The percentage of ALP activity relative to that of starved control cells is shown. The values represent averages of four independent experiments, with the SD as error bars. (B) Chemical genetic analysis of ATG1 using SC-grown cells. Cells (TN124–1C, atg1-1 pho8_Δ_60) harboring the indicated ATG1 plasmids were grown in SC-Ura medium. Thecells were treated with 1-NA-PP1, incubated in SD(-N) medium, and analyzed as described in A. The values (values of the starved control cells were normalized to 100%) represent the mean ALP activities from four independent experiments. The results from YEPD-grown cells (shown in A) are also shown. (C) Autophosphorylation activity of Atg1M102A is inhibited by 20 μM 1-NA-PP1. Cell extract of the HAAtg1M102A mutant cells before (left) and after (right) nitrogen starvation for 4 h was immunoprecipitated with anti-HA antibody, and the resultant immunocomplex was assayed for autophosphorylation activity in the presence of or absence of 20 μM 1-NA-PP1. (D) Atg1 kinase activity is impaired in _atg17_C24R mutant cells. Wild-type (YYK422), atg17C24R mutant (YYK467), or _atg17_Δ mutant (YYK416) cells were treated with rapamycin (0.2 μg/ml, 30 min) and subjected to Atg1 kinase assay. (E) Atg1 kinase activity in phosphorylating itself is increased in starvation condition. HAAtg1WT, immunoprecipitated as described in C, was subjected to autophosphorylation assay (top), to protein kinase assay using 4 μg of recombinant Atg1 protein as a substrate (middle), and to protein kinase assay using 4 μg of myelin basic protein (MBP, bottom).
Similar articles
- Dual role of Atg1 in regulation of autophagy-specific PAS assembly in Saccharomyces cerevisiae.
Cheong H, Klionsky DJ. Cheong H, et al. Autophagy. 2008 Jul;4(5):724-6. doi: 10.4161/auto.6375. Epub 2008 Jun 2. Autophagy. 2008. PMID: 18552550 - Molecular interactions of the Saccharomyces cerevisiae Atg1 complex provide insights into assembly and regulatory mechanisms.
Chew LH, Lu S, Liu X, Li FK, Yu AY, Klionsky DJ, Dong MQ, Yip CK. Chew LH, et al. Autophagy. 2015;11(6):891-905. doi: 10.1080/15548627.2015.1040972. Autophagy. 2015. PMID: 25998554 Free PMC article. - The Intrinsically Disordered Protein Atg13 Mediates Supramolecular Assembly of Autophagy Initiation Complexes.
Yamamoto H, Fujioka Y, Suzuki SW, Noshiro D, Suzuki H, Kondo-Kakuta C, Kimura Y, Hirano H, Ando T, Noda NN, Ohsumi Y. Yamamoto H, et al. Dev Cell. 2016 Jul 11;38(1):86-99. doi: 10.1016/j.devcel.2016.06.015. Dev Cell. 2016. PMID: 27404361 - ATG13: just a companion, or an executor of the autophagic program?
Alers S, Wesselborg S, Stork B. Alers S, et al. Autophagy. 2014 Jun;10(6):944-56. doi: 10.4161/auto.28987. Autophagy. 2014. PMID: 24879146 Free PMC article. Review. - [Molecular mechanisms of autophagy in yeast].
Nakatogawa H, Ohsumi Y. Nakatogawa H, et al. Tanpakushitsu Kakusan Koso. 2008 Dec;53(16 Suppl):2099-105. Tanpakushitsu Kakusan Koso. 2008. PMID: 21038592 Review. Japanese. No abstract available.
Cited by
- Autophagy, a process within reperfusion injury: an update.
Thapalia BA, Zhou Z, Lin X. Thapalia BA, et al. Int J Clin Exp Pathol. 2014 Dec 1;7(12):8322-41. eCollection 2014. Int J Clin Exp Pathol. 2014. PMID: 25674198 Free PMC article. Review. - An in vivo detection system for transient and low-abundant protein interactions and their kinetics in budding yeast.
Brezovich A, Schuschnig M, Ammerer G, Kraft C. Brezovich A, et al. Yeast. 2015 Mar;32(3):355-65. doi: 10.1002/yea.3063. Epub 2015 Feb 10. Yeast. 2015. PMID: 25582094 Free PMC article. - AMPK-dependent phosphorylation of ULK1 regulates ATG9 localization.
Mack HI, Zheng B, Asara JM, Thomas SM. Mack HI, et al. Autophagy. 2012 Aug;8(8):1197-214. doi: 10.4161/auto.20586. Epub 2012 Aug 1. Autophagy. 2012. PMID: 22932492 Free PMC article. - Network organization of the human autophagy system.
Behrends C, Sowa ME, Gygi SP, Harper JW. Behrends C, et al. Nature. 2010 Jul 1;466(7302):68-76. doi: 10.1038/nature09204. Epub 2010 Jun 20. Nature. 2010. PMID: 20562859 Free PMC article. - Direct induction of autophagy by Atg1 inhibits cell growth and induces apoptotic cell death.
Scott RC, Juhász G, Neufeld TP. Scott RC, et al. Curr Biol. 2007 Jan 9;17(1):1-11. doi: 10.1016/j.cub.2006.10.053. Curr Biol. 2007. PMID: 17208179 Free PMC article.
References
- Bishop, A. C., Kung, C.-y., Shah, K., Witucki, L., Shokat, K. M., and Liu, Y. (1999). Generation of monospecific nanomolar tyrosine kinase inhibitors via a chemical genetic approach. J. Am. Chem. Soc. 121, 627-631.
- Dieffenbach, C. W., and Dveksler, G. S. (1995). PCR Primer: A Laboratory Manual, Plainview, NY: Cold Spring Harbor Laboratory Press.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases