A high-resolution linkage-disequilibrium map of the human major histocompatibility complex and first generation of tag single-nucleotide polymorphisms - PubMed (original) (raw)
doi: 10.1086/429393. Epub 2005 Mar 1.
Emily C Walsh, Xiayi Ke, Marcos Delgado, Mark Griffiths, Sarah Hunt, Jonathan Morrison, Pamela Whittaker, Eric S Lander, Lon R Cardon, David R Bentley, John D Rioux, Stephan Beck, Panos Deloukas
Affiliations
- PMID: 15747258
- PMCID: PMC1199300
- DOI: 10.1086/429393
A high-resolution linkage-disequilibrium map of the human major histocompatibility complex and first generation of tag single-nucleotide polymorphisms
Marcos M Miretti et al. Am J Hum Genet. 2005 Apr.
Abstract
Autoimmune, inflammatory, and infectious diseases present a major burden to human health and are frequently associated with loci in the human major histocompatibility complex (MHC). Here, we report a high-resolution (1.9 kb) linkage-disequilibrium (LD) map of a 4.46-Mb fragment containing the MHC in U.S. pedigrees with northern and western European ancestry collected by the Centre d'Etude du Polymorphisme Humain (CEPH) and the first generation of haplotype tag single-nucleotide polymorphisms (tagSNPs) that provide up to a fivefold increase in genotyping efficiency for all future MHC-linked disease-association studies. The data confirm previously identified recombination hotspots in the class II region and allow the prediction of numerous novel hotspots in the class I and class III regions. The region of longest LD maps outside the classic MHC to the extended class I region spanning the MHC-linked olfactory-receptor gene cluster. The extended haplotype homozygosity analysis for recent positive selection shows that all 14 outlying haplotype variants map to a single extended haplotype, which most commonly bears HLA-DRB1*1501. The SNP data, haplotype blocks, and tagSNPs analysis reported here have been entered into a multidimensional Web-based database (GLOVAR), where they can be accessed and viewed in the context of relevant genome annotation. This LD map allowed us to give coordinates for the extremely variable LD structure underlying the MHC.
Figures
Figure 1
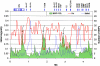
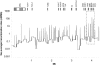
Low-heterozygosity regions in the MHC. The observed heterozygosity (“Ho,” red line) averaged in 50-kb windows is plotted with the number of loci with MAF <5% (green area). SNPs showing MAF <5% are not randomly distributed across the analyzed 4.459-Mb region. Specifically, three regions (black arrows) presented loss of heterozygosity; most SNPs are monomorphic in this population. These fragments contain OR2H1 and MAS1L; DHX16, NRM, MDC1 TUBB, and FLOT1; BAT5, LY6GD, C6orf25, DDAH2, C6orf26, and VARS2, respectively. The uneven distribution of the 770 SNPs that failed to generate genotyping data is represented by the blue line (“failed,” number of SNPs in 50-kb windows). Typing failures are concentrated mainly in three genomic regions (blue peaks), including HLA-A, HLA-B, HLA-C, HLA-DQB1, and HLA-DRB1, which suggests that the highly polymorphic nature of these genes might be responsible for the failure.
Figure 2
SW plot of average
_r_2
across the MHC. Average
_r_2
was calculated from 25 kb to 250 kb in 500-kb SWs, with 50-kb increments between windows. The SW plot captures trends of LD (
_r_2
) by averaging
_r_2
values between a given marker and all the SNPs up to 250 kb. Avoiding pairwise comparisons with surrounding markers (25 kb each side) excludes the raising effect of closely linked loci on LD. MHC extended class I, class I, class III, and class II regions (blue, yellow, orange, and green, respectively) present comparatively distinct variation patterns of long-range LD, which is reflected in the haplotype-block analysis and interferes in the SNP-tagging process.
Figure 3
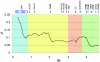
Decay of LD as a function of distance. The decay rates represented by average
_r_2
(A) and D′ (B) for all marker pairs separated by distance S (
_S_=10
kb, 20 kb, 30 kb,…500 kb). Line colors represent different MHC subregions and chromosome 20 genotyping data from CEPH families (Ke et al. 2004_b_). Whereas LD decay values averaged across the whole MHC region are comparable with the chromosome 20 LD decay, the extended class I and classical class II regions show distinct slopes consistent with the underlying LD structure observed in fig. 2.
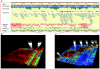
Figure 4
LD structure across the MHC. A, Distribution of haplotype blocks across the MHC region, as viewed in the GLOVAR genome browser. Haplotype blocks, according to criteria of Gabriel et al. (2002) implemented by Haploview 2.05 (Haploview Web site), are represented by red bars. Each bar corresponds to an individual haplotype block comprising a number of SNPs (red marks), which are located according their map position. This enables an accurate interpretation of the LD-block distribution, size, and gaps in the context of additional genomic features, such as gene annotation, SNP density, and physical distance. The distribution of tagSNPs selected in this work is generated in the GLOVAR genome browser and is indicated by a green track under the haplotype blocks. B, High-resolution view of 720 kb of the extended MHC class I region, as represented by GOLDsurfer 3D view of D′ values (Pettersson et al. 2004). This region contains a large cluster of olfactory-receptor genes in high LD (540 kb), interrupted by a single recombination hotspot between OR12D3 and OR12D2. This long-range LD region includes 13 contiguous haplotype blocks, according to the criteria of Gabriel et al. (2002) (see corresponding inset in panel A). C, View of the LD structure (D′ values) within the MHC class II region in which experimental evidence for recombination hotspots have been described elsewhere (Cullen et al. ; Jeffreys et al. 2001). High-LD areas (red blocks) are separated by recombination hotspots. The first three LD breaks correspond to recombination hotspots mapped at TAP2 and HLA-DMB and between BRD2 and HLA-DOA (Cullen et al. ; Jeffreys et al. 2001). Another LD break is visualized between HLA-DOA and HLA-DPA1.
Figure 5
Recombination-rate variation across the MHC. Recombination rates estimated from population-genetic data are far from being uniform; their distribution fluctuates considerably in both, by scale (cM/Mb) and by map position. Recombination hotspots are represented by peaks 10 times higher than the local background level of recombination. Peaks enclosed in the inset correspond to hotspots identified by sperm typing (Jeffreys et al. 2001) located at or near TAP2, HLA-DMB, BRD2, and HLA-DOA, observable as LD breaks in figure 4_C_ . The recombination hotspot between OR12D3 and OR12D2 in the olfactory-receptor gene cluster (“ORs”), inferred from population genetic data, correlates perfectly with the LD break visible in figure 4_B_ . Note the presence of two coldspots showing recombination rates 10 times lower than the local background level of recombination.
Figure 6
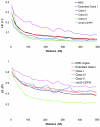
EHH outliers at 0.3 cM and 0.25 cM. A, EHH by frequency plot at 0.3cM, indicating the eight outliers at that distance. B, EHH by frequency plot at 0.25cM, indicating six outliers at that distance. C, Physical mapping of outlying variants. A subset of genes in the region is shown for reference. Haplotype variants with extended LD are indicated in orange and with numbers that correlate to their position in the EHH by frequency plots in panels A and B.
Figure 7
Representative EHH outlier. A, EHH by frequency plot at 0.3 cM distance, indicating one of the eight variants at that an outlier with distance that is >4.5 SD in its frequency bin and per its
frequency_×_EHH
statistic. This variant is indicated in figure 6 (panel 9) and is part of a block slightly centromeric to the DQB1 gene. B, Haplotype structure of the block containing this haplotype variant. The outlier is the 37% allele (“GGGT”) indicated in orange in all remaining plots (arrow). C, EHH by distance (cM) plot of all variants in the block. The arrow indicates the region of extended LD for the orange haplotype. D, Haplotype bifurcation plots of all variants in the block. The arrow indicates the region of interest for the orange haplotype.
Similar articles
- Linkage disequilibrium and haplotype blocks in the MHC vary in an HLA haplotype specific manner assessed mainly by DRB1*03 and DRB1*04 haplotypes.
Blomhoff A, Olsson M, Johansson S, Akselsen HE, Pociot F, Nerup J, Kockum I, Cambon-Thomsen A, Thorsby E, Undlien DE, Lie BA. Blomhoff A, et al. Genes Immun. 2006 Mar;7(2):130-40. doi: 10.1038/sj.gene.6364272. Genes Immun. 2006. PMID: 16395395 - Similarity in recombination rate and linkage disequilibrium at CYP2C and CYP2D cytochrome P450 gene regions among Europeans indicates signs of selection and no advantage of using tagSNPs in population isolates.
Pimenoff VN, Laval G, Comas D, Palo JU, Gut I, Cann H, Excoffier L, Sajantila A. Pimenoff VN, et al. Pharmacogenet Genomics. 2012 Dec;22(12):846-57. doi: 10.1097/FPC.0b013e32835a3a6d. Pharmacogenet Genomics. 2012. PMID: 23089684 - Short tandem repeat (STR) haplotypes in HLA: an integrated 50-kb STR/linkage disequilibrium/gene map between the RING3 and HLA-B genes and identification of STR haplotype diversification in the class III region.
Vorechovsky I, Kralovicova J, Laycock MD, Webster AD, Marsh SG, Madrigal A, Hammarström L. Vorechovsky I, et al. Eur J Hum Genet. 2001 Aug;9(8):590-8. doi: 10.1038/sj.ejhg.5200688. Eur J Hum Genet. 2001. PMID: 11528504 - Recombination within the human MHC.
Carrington M. Carrington M. Immunol Rev. 1999 Feb;167:245-56. doi: 10.1111/j.1600-065x.1999.tb01397.x. Immunol Rev. 1999. PMID: 10319266 Review. - The human Major Histocompatibility Complex as a paradigm in genomics research.
Vandiedonck C, Knight JC. Vandiedonck C, et al. Brief Funct Genomic Proteomic. 2009 Sep;8(5):379-94. doi: 10.1093/bfgp/elp010. Epub 2009 May 25. Brief Funct Genomic Proteomic. 2009. PMID: 19468039 Free PMC article. Review.
Cited by
- Immunogenomics: molecular hide and seek.
Miretti MM, Beck S. Miretti MM, et al. Hum Genomics. 2006 Jan;2(4):244-51. doi: 10.1186/1479-7364-2-4-244. Hum Genomics. 2006. PMID: 16460649 Free PMC article. Review. - The HLA Region and Autoimmune Disease: Associations and Mechanisms of Action.
Gough SC, Simmonds MJ. Gough SC, et al. Curr Genomics. 2007 Nov;8(7):453-65. doi: 10.2174/138920207783591690. Curr Genomics. 2007. PMID: 19412418 Free PMC article. - The architecture of long-range haplotypes shared within and across populations.
Gusev A, Palamara PF, Aponte G, Zhuang Z, Darvasi A, Gregersen P, Pe'er I. Gusev A, et al. Mol Biol Evol. 2012 Feb;29(2):473-86. doi: 10.1093/molbev/msr133. Epub 2011 Oct 6. Mol Biol Evol. 2012. PMID: 21984068 Free PMC article. - Implications of inter-population linkage disequilibrium patterns on the approach to a disease association study in the human MHC class III.
Hanchard N, Diakite M, Koch O, Keating B, Pinder M, Jallow M, Sisay-Joof F, Nijnik A, Wilson J, Udalova I, Kwiatkowski D, Rockett K. Hanchard N, et al. Immunogenetics. 2006 Jun;58(5-6):465-70. doi: 10.1007/s00251-006-0118-1. Epub 2006 Apr 28. Immunogenetics. 2006. PMID: 16738941 - Whole Genome DNA and RNA Sequencing of Whole Blood Elucidates the Genetic Architecture of Gene Expression Underlying a Wide Range of Diseases.
Liu C, Joehanes R, Ma J, Wang Y, Sun X, Keshawarz A, Sooda M, Huan T, Hwang SJ, Bui H, Tejada B, Munson PJ, Cumhur D, Heard-Costa NL, Pitsillides AN, Peloso GM, Feolo M, Sharopova N, Vasan RS, Levy D. Liu C, et al. Res Sq [Preprint]. 2022 May 31:rs.3.rs-1598646. doi: 10.21203/rs.3.rs-1598646/v1. Res Sq. 2022. PMID: 35664994 Free PMC article. Updated. Preprint.
References
Electronic-Database Information
- Center for Statistical Genetics, http://www.sph.umich.edu/csg/abecasis/PedStats/ (for PEDSTATS)
- dbSNP Home Page, http://www.ncbi.nlm.nih.gov/SNP/index.html
- GLOVAR Genome Browser, http://www.glovar.org/Homo_sapiens/
- Human Chromosome 6 Project Overview, http://www.sanger.ac.uk/HGP/Chr6/
References
- Abecasis GR, Cherny SS, Cookson WO, Cardon LR (2002) MERLIN—rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet 30:97–101 - PubMed
- Abecasis GR, Cookson WOC (2000) GOLD—graphical overview of linkage disequilibrium. Bioinformatics 16:182–183 - PubMed
- Ahmad T, Neville M, Marshall SE, Armuzzi A, Mulcahy-Hawes K, Crawshaw J, Sato H, Ling K-L, Barnardo M, Goldthorpe S, Walton R, Bunce M, Jewell DP, Welsh KI (2003) Haplotype-specific linkage disequilibrium patterns define the genetic topography of the human MHC. Hum Mol Genet 12:647–656 - PubMed
- Barcellos LF, Oksenberg JR, Begovich AB, Martin ER, Schmidt S, Vittinghoff E, Goodin DS, Pelletier D, Lincoln RR, Bucher P, Swerdlin A, Perick-Vance MA, Haines JL, Hauser SL, for the Multiple Sclerosis Genetics Group (2003) HLA-DR2 dose effect on susceptibility to multiple sclerosis and influence on disease course. Am J Hum Genet 72:710–716 - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous