From genome to antivirals: SARS as a test tube - PubMed (original) (raw)
Review
From genome to antivirals: SARS as a test tube
Yossef Kliger et al. Drug Discov Today. 2005.
Abstract
The severe acute respiratory syndrome (SARS) epidemic brought into the spotlight the need for rapid development of effective anti-viral drugs against newly emerging viruses. Researchers have leveraged the 20-year battle against AIDS into a variety of possible treatments for SARS. Most prominently, based solely on viral genome information, silencers of viral genes, viral-enzyme blockers and viral-entry inhibitors were suggested as potential therapeutic agents for SARS. In particular, inhibitors of viral entry, comprising therapeutic peptides, were based on the recently launched anti-HIV drug enfuvirtide. This could represent one of the most direct routes from genome sequencing to the discovery of antiviral drugs.
Figures
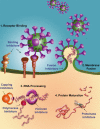
FIGURE 1
The SARS-CoV life cycle is vulnerable to therapeutic intervention in several places. (1) Virus binding to cellular receptors. Outside the cell, blocking the interaction of SARS-CoV with the cellular receptor will prevent the virus from attaching to host cells. Co-receptor antagonists will prevent the initiation of the next step. (2) Membrane fusion of the virion with the host. Fusion inhibitors will block merging of the viral membrane with the host cell membrane. (3) Viral RNA processing. Within the cell, transcription and multiplication of the viral RNA can be blocked by polymerase and helicase inhibitors. Translation of the viral proteins might be inhibited by blocking mRNA capping. (4) Protein maturation. Viral proteins will not mature in the presence of protease inhibitors, rendering them useless.
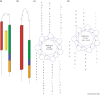
FIGURE 2
Similarity between the fusion proteins of HIV-1 and SARS-CoV. Schematic illustration of (a) HIV-1 gp41 and (b) the equivalent S2 protein from the SARS-CoV. A Leucine/Isoleucine heptad repeat adjacent to the N-terminus of both proteins appears in red. The C-HR is in green. Cysteine residues (purple) confining a loop structure are located between the two heptad repeats. An aromatic residues-rich motif is marked blue, and the transmembrane segment is in orange. A peptide corresponding to the C-HR, which acts as potent inhibitor of HIV-1 entry into the cell, appears in yellow. The helical wheel is a top view of a single strand of a coiled coil. In the wheel projection of the N-HR (c) and C-HR (d) of SARS-CoV S2 protein, each of the seven positions (a-g) corresponds to the location of an amino acid residue that makes up the coiled coil. The arrows between the seven positions indicate the relative locations of adjacent residues in an amino acid subsequence. The helical wheel demonstrates how a potential antiviral drug can be discovered based solely on sequence information.
FIGURE 3
A time line comparing key achievements in AIDS and SARS research. Effective international collaborations and technological advances greatly accelerated the understanding of viral diseases. It is anticipated that these research achievements will also lead to faster drug discovery and development.
Similar articles
- Recent developments in the virology and antiviral research of severe acute respiratory syndrome coronavirus.
Yeung KS, Meanwell NA. Yeung KS, et al. Infect Disord Drug Targets. 2007 Mar;7(1):29-41. doi: 10.2174/187152607780090739. Infect Disord Drug Targets. 2007. PMID: 17346209 Review. - Potential targets for anti-SARS drugs in the structural proteins from SARS related coronavirus.
Wu G, Yan S. Wu G, et al. Peptides. 2004 Jun;25(6):901-8. doi: 10.1016/j.peptides.2004.03.002. Peptides. 2004. PMID: 15203235 Free PMC article. - Recent patents on treatment of severe acute respiratory syndrome (SARS).
Zhai S, Liu W, Yan B. Zhai S, et al. Recent Pat Antiinfect Drug Discov. 2007 Jan;2(1):1-10. doi: 10.2174/157489107779561698. Recent Pat Antiinfect Drug Discov. 2007. PMID: 18221160 Review. - Potential antivirals and antiviral strategies against SARS coronavirus infections.
De Clercq E. De Clercq E. Expert Rev Anti Infect Ther. 2006 Apr;4(2):291-302. doi: 10.1586/14787210.4.2.291. Expert Rev Anti Infect Ther. 2006. PMID: 16597209 Free PMC article. Review. - Infectious diseases. One year after outbreak, SARS virus yields some secrets.
Enserink M. Enserink M. Science. 2004 May 21;304(5674):1097. doi: 10.1126/science.304.5674.1097. Science. 2004. PMID: 15155925 No abstract available.
Cited by
- Investigation of the pharmacophore space of Severe Acute Respiratory Syndrome coronavirus (SARS-CoV) NTPase/helicase by dihydroxychromone derivatives.
Lee C, Lee JM, Lee NR, Kim DE, Jeong YJ, Chong Y. Lee C, et al. Bioorg Med Chem Lett. 2009 Aug 15;19(16):4538-41. doi: 10.1016/j.bmcl.2009.07.009. Epub 2009 Jul 9. Bioorg Med Chem Lett. 2009. PMID: 19625187 Free PMC article. - Three dimensional model of severe acute respiratory syndrome coronavirus helicase ATPase catalytic domain and molecular design of severe acute respiratory syndrome coronavirus helicase inhibitors.
Hoffmann M, Eitner K, von Grotthuss M, Rychlewski L, Banachowicz E, Grabarkiewicz T, Szkoda T, Kolinski A. Hoffmann M, et al. J Comput Aided Mol Des. 2006 May;20(5):305-19. doi: 10.1007/s10822-006-9057-z. Epub 2006 Sep 14. J Comput Aided Mol Des. 2006. PMID: 16972168 Free PMC article. - Traditional Chinese medicine herbal extracts of Cibotium barometz, Gentiana scabra, Dioscorea batatas, Cassia tora, and Taxillus chinensis inhibit SARS-CoV replication.
Wen CC, Shyur LF, Jan JT, Liang PH, Kuo CJ, Arulselvan P, Wu JB, Kuo SC, Yang NS. Wen CC, et al. J Tradit Complement Med. 2011 Oct;1(1):41-50. doi: 10.1016/s2225-4110(16)30055-4. J Tradit Complement Med. 2011. PMID: 24716104 Free PMC article. - The University of Washington Health Sciences Library BioCommons: an evolving Northwest biomedical research information support infrastructure.
Minie M, Bowers S, Tarczy-Hornoch P, Roberts E, James RA, Rambo N, Fuller S. Minie M, et al. J Med Libr Assoc. 2006 Jul;94(3):321-9. J Med Libr Assoc. 2006. PMID: 16888667 Free PMC article. - SARS coronavirus entry into host cells through a novel clathrin- and caveolae-independent endocytic pathway.
Wang H, Yang P, Liu K, Guo F, Zhang Y, Zhang G, Jiang C. Wang H, et al. Cell Res. 2008 Feb;18(2):290-301. doi: 10.1038/cr.2008.15. Cell Res. 2008. PMID: 18227861 Free PMC article.
References
- Ksiazek T.G. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1953–1966. - PubMed
- Drosten C. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1967–1976. - PubMed
- Marra M.A. The Genome sequence of the SARS-associated coronavirus. Science. 2003;300:1399–1404. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous