HISTONE DEACETYLASE19 is involved in jasmonic acid and ethylene signaling of pathogen response in Arabidopsis - PubMed (original) (raw)
HISTONE DEACETYLASE19 is involved in jasmonic acid and ethylene signaling of pathogen response in Arabidopsis
Changhe Zhou et al. Plant Cell. 2005 Apr.
Abstract
Histone acetylation is modulated through the action of histone acetyltransferases and deacetylases, which play key roles in the regulation of eukaryotic gene expression. Previously, we have identified a yeast histone deacetylase REDUCED POTASSIUM DEPENDENCY3 (RPD3) homolog, HISTONE DEACETYLASE19 (HDA19) (AtRPD3A), in Arabidopsis thaliana. Here, we report further study of the expression and function of HDA19. Analysis of Arabidopsis plants containing the HDA19:beta-glucuronidase fusion gene revealed that HDA19 was expressed throughout the life of the plant and in most plant organs examined. In addition, the expression of HDA19 was induced by wounding, the pathogen Alternaria brassicicola, and the plant hormones jasmonic acid and ethylene. Using green fluorescent protein fusion, we demonstrated that HDA19 accumulated in the nuclei of Arabidopsis cells. Overexpression of HDA19 in 35S:HDA19 plants decreased histone acetylation levels, whereas downregulation of HDA19 in HDA19-RNA interference (RNAi) plants increased histone acetylation levels. In comparison with wild-type plants, 35S:HDA19 transgenic plants had increased expression of ETHYLENE RESPONSE FACTOR1 and were more resistant to the pathogen A. brassicicola. The expression of jasmonic acid and ethylene regulated PATHOGENESIS-RELATED genes, Basic Chitinase and beta-1,3-Glucanase, was upregulated in 35S:HDA19 plants but downregulated in HDA19-RNAi plants. Our studies provide evidence that HDA19 may regulate gene expression involved in jasmonic acid and ethylene signaling of pathogen response in Arabidopsis.
Figures
Figure 1.
GUS Activity in HDA19:GUS Plants. GUS staining patterns of a seedling (A), leaf (B), stem (C), flower (D), stigma (E), anther (F), siliques (G), and seeds (H) from HDA19:GUS plants. GUS staining in HDA19:GUS leaves without treatment (I), treated with 1 mM methyl-jasmonate (JA) (J), 1 mM ACC (an ethylene precursor) (K), or 10 mM MgCl2 (L), infected with A. brassicicola for 24 h (M), and after cutting (N).
Figure 2.
RT-PCR Analysis of Expression of HDAC Genes. Total RNA for RT-PCR analysis was isolated from leaf tissues of Arabidopsis plants treated without (control) or with 1 mM JA and 1 mM ACC (an ethylene precursor) for 6 h. Ubiquitin (UBQ) is shown as an internal control.
Figure 3.
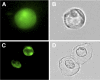
Subcellular Localization of HDA19. Protoplasts were isolated from the leaves of 35S:GFP ([A] and [B]) and 35S:HDA19-GFP ([C] and [D]) transgenic Arabidopsis plants. GFP fluorescence was examined by fluorescence microscopy under UV light ([A] and [C]) and white light ([B] and [D]).
Figure 4.
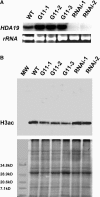
Levels of Tetra-Acetylated H3 in the 35S:HDA19 and _HDA19_-RNAi Lines. (A) RNA gel blot analysis of HDA19 expression in wild-type, 35S:HDA19 (G11-1, G11-2, and G11-3), and _HDA19-_RNAi (RNAi-1 and RNAi-2) plants. Twenty micrograms of total RNA were probed with HDA19. Ethidium bromide–stained rRNA are shown to illustrate the gel loading. (B) Protein gel blot analysis detecting tetra-acetylated H3 (H3ac) (top panel) using α-H3ac antibodies on protein extracts from wild-type, 35S:HDA19 (G11-1, G11-2, and G11-3), and HDA19-RNAi (RNAi-1 and RNAi-2) plants. Bottom panel, Coomassie blue staining shows equal protein loading. MW, molecular weight marker.
Figure 5.
Phenotypic Abnormalities in 35S:HDA19 Plants. (A) to (C) The 35S:HDA19 transgenic seedlings from the T2 line showed aberrant cotyledons and lacked shoot and root development ([B] and [C]) compared with a wild-type seedling (A). (D) to (F) The 35S:HDA19 transgenic plants from the T2 line with branching leaf (E) and narrow leaf (F) compared with a normal leaf (D). (G) and (H) A 6-week-old 35S:HDA19 plant (H) from the T2 line showed delayed flowering when compared with a 6-week-old wild-type plant (G). (I) and (J) A stem from a 35S:HDA19 transgenic plant of the T2 line with stunted siliques (I) and a wild-type stem with full silique elongation (J).
Figure 6.
Gene Expression in the 35S:HDA19 and _HDA19_-RNAi Plants. (A) RT-PCR analysis of ERF1, BGL, and CHI-B expression in wild-type (1), 35S:HDA19 (2 and 3), and _HDA19-_RNAi (4 and 5) transgenic lines. (B) RT-PCR analysis of VSP2 and RNS1 expression in wild-type (1), 35S:HDA19 (2 and 3), and _HDA19-_RNAi (4 and 5) transgenic lines. Total RNA for RT-PCR analysis was isolated from leaf tissues of Arabidopsis plants. Ubiquitin (UBQ) is shown as an internal control.
Figure 7.
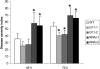
Resistance to A. brassicicola by HDA19 Overexpression. Graphical representation of disease symptoms in wild-type, 35S:HDA19 (G11-1 and G11-2), and _HDA19-_RNAi (RNAi-1 and RNAi-2) transgenic lines. Disease severity index was calculated 48 and 72 h after inoculation with A. brassicicola. Disease severity index was calculated based on the degree of symptom severity as measured by leaf necrosis area: 0, no symptom; 1, 1 to 5% necrosis areas; 2, 5 to 10% necrosis areas; 3, 10 to 25% necrosis areas; 4, >25% necrosis areas. The leaf necrosis severity ratings were summed for 60 to 80 leaves of 15 to 25 plants per genotype and divided by the number of leaves rated times the maximum possible rating to given the final disease severity index. Asterisks mark values that are significantly different from the wild type (χ2 test, P < 0.05). The experiment was repeated three times with similar results.
Similar articles
- Ethylene modulates the role of NONEXPRESSOR OF PATHOGENESIS-RELATED GENES1 in cross talk between salicylate and jasmonate signaling.
Leon-Reyes A, Spoel SH, De Lange ES, Abe H, Kobayashi M, Tsuda S, Millenaar FF, Welschen RA, Ritsema T, Pieterse CM. Leon-Reyes A, et al. Plant Physiol. 2009 Apr;149(4):1797-809. doi: 10.1104/pp.108.133926. Epub 2009 Jan 28. Plant Physiol. 2009. PMID: 19176718 Free PMC article. - The Negative Regulators of the Basal Defence WRKY7, WRKY11 and WRKY17 Modulate the Jasmonic Acid Pathway and an Alternative Splicing Regulatory Network in Response to Pseudomonas syringae in Arabidopsis thaliana.
Fuenzalida-Valdivia I, Herrera-Vásquez A, Gangas MV, Sáez-Vásquez J, Álvarez JM, Meneses C, Blanco-Herrera F. Fuenzalida-Valdivia I, et al. Mol Plant Pathol. 2024 Dec;25(12):e70044. doi: 10.1111/mpp.70044. Mol Plant Pathol. 2024. PMID: 39717006 Free PMC article. - The epiphytic fungus Pseudozyma aphidis induces jasmonic acid- and salicylic acid/nonexpressor of PR1-independent local and systemic resistance.
Buxdorf K, Rahat I, Gafni A, Levy M. Buxdorf K, et al. Plant Physiol. 2013 Apr;161(4):2014-22. doi: 10.1104/pp.112.212969. Epub 2013 Feb 6. Plant Physiol. 2013. PMID: 23388119 Free PMC article. - Depressing time: Waiting, melancholia, and the psychoanalytic practice of care.
Salisbury L, Baraitser L. Salisbury L, et al. In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. PMID: 36137063 Free Books & Documents. Review. - Impact of residual disease as a prognostic factor for survival in women with advanced epithelial ovarian cancer after primary surgery.
Bryant A, Hiu S, Kunonga PT, Gajjar K, Craig D, Vale L, Winter-Roach BA, Elattar A, Naik R. Bryant A, et al. Cochrane Database Syst Rev. 2022 Sep 26;9(9):CD015048. doi: 10.1002/14651858.CD015048.pub2. Cochrane Database Syst Rev. 2022. PMID: 36161421 Free PMC article. Review.
Cited by
- HISTONE DEACETYLASE6 Controls Gene Expression Patterning and DNA Methylation-Independent Euchromatic Silencing.
Hristova E, Fal K, Klemme L, Windels D, Bucher E. Hristova E, et al. Plant Physiol. 2015 Aug;168(4):1298-308. doi: 10.1104/pp.15.00177. Epub 2015 Apr 27. Plant Physiol. 2015. PMID: 25918117 Free PMC article. - Arabidopsis thaliana histone deacetylase 1 (AtHD1) is localized in euchromatic regions and demonstrates histone deacetylase activity in vitro.
Fong PM, Tian L, Chen ZJ. Fong PM, et al. Cell Res. 2006 May;16(5):479-88. doi: 10.1038/sj.cr.7310059. Cell Res. 2006. PMID: 16699543 Free PMC article. - Chromatin enrichment for proteomics in plants (ChEP-P) implicates the histone reader ALFIN-LIKE 6 in jasmonate signalling.
Vélez-Bermúdez IC, Schmidt W. Vélez-Bermúdez IC, et al. BMC Genomics. 2021 Nov 22;22(1):845. doi: 10.1186/s12864-021-08160-6. BMC Genomics. 2021. PMID: 34809577 Free PMC article. - HISTONE MONOUBIQUITINATION1 interacts with a subunit of the mediator complex and regulates defense against necrotrophic fungal pathogens in Arabidopsis.
Dhawan R, Luo H, Foerster AM, Abuqamar S, Du HN, Briggs SD, Mittelsten Scheid O, Mengiste T. Dhawan R, et al. Plant Cell. 2009 Mar;21(3):1000-19. doi: 10.1105/tpc.108.062364. Epub 2009 Mar 13. Plant Cell. 2009. PMID: 19286969 Free PMC article. - Phylogenetic analysis, subcellular localization, and expression patterns of RPD3/HDA1 family histone deacetylases in plants.
Alinsug MV, Yu CW, Wu K. Alinsug MV, et al. BMC Plant Biol. 2009 Mar 28;9:37. doi: 10.1186/1471-2229-9-37. BMC Plant Biol. 2009. PMID: 19327164 Free PMC article.
References
- Aravind, L., and Koonin, E.V. (1998). Second family of histone deacetylases. Science 280, 1167a.
- Ausin, I., Alonso-Blanco, C., Jarillo, J.A., Ruiz-Garcia, L., and Martinez-Zapater, J.M. (2004). Regulation of flowering time by FVE, a retinoblastoma-associated protein. Nat. Genet. 36, 162–166. - PubMed
- Berger, S.L. (2002). Histone modification in transcriptional regulation. Curr. Opin. Genet. Dev. 12, 142–148. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases