Transcriptional network underlying Caenorhabditis elegans vulval development - PubMed (original) (raw)
Transcriptional network underlying Caenorhabditis elegans vulval development
Takao Inoue et al. Proc Natl Acad Sci U S A. 2005.
Abstract
The vulval development of Caenorhabditis elegans provides an opportunity to investigate genetic networks that control gene expression during organogenesis. During the fourth larval stage (L4), seven vulval cell types are produced, each of which executes a distinct gene expression program. We analyze how the expression of cell-type-specific genes is regulated. Ras and Wnt signaling pathways play major roles in generating the spatial pattern of cell types and regulate gene expression through a network of transcription factors. One transcription factor (lin-29) primarily controls the temporal expression pattern. Other transcription factors (lin-11, cog-1, and egl-38) act in combination to control cell-type-specific gene expression. The complexity of the network arises in part because of the dynamic nature of gene expression, in part because of the presence of seven cell types, and also because there are multiple regulatory paths for gene expression within each cell type.
Figures
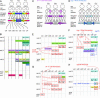
Fig. 1.
The pattern of gene expression during late stages of vulval development. (A) An overview of vulval development. Lineal origins of 22 vulval nuclei are indicated. “ABCDEFFEDCBA” refer to vulval cell types vulA, vulB1, vulB2, vulC, vulD, vulE, and vulF. vulB is the only case in which a single VPC granddaughter gives rise to two cell types (8). Vulval cell nuclei at each stage are positioned as indicated (left, anterior; right, posterior). (B) A summary of cell-type specificity and timing of expression in the wild type (Materials and Methods) (, , –, –26). Boxes indicate stages at which gene expression is activated. The vertical order of events within each time block is arbitrary. For egl-17, vulE/vulF expression begins in P6.p (early L3) and persists in their descendants (vulE and vulF) until turned off in the early L4 stage. This inactivation, which is regulated by lin-29 and lin-11, is indicated by the box marked “egl-17 OFF.” ceh-2 is expressed at a higher level in vulB1 compared with vulB2. (C) Expression pattern of lin-11. The diagrammed pattern is based on the lin-11::gfp transgene syIs80 (26). (D) Expression pattern of cog-1. The pattern is based on the cog-1::gfp transgene syIs63 (13). (E) The altered pattern of gene expression in lin-29 mutants (9, 23) (Tables 1 and 2). White boxes with the red outlines indicate loss of expression and loss of egl-17 down-regulation in the lin-29 mutant. lin-29 appears to regulate events that occur during the mid-L4 to the late L4 stage. (F) The altered pattern of gene expression in cog-1 mutants (Table 3 and Fig. 2). Arrows are drawn with the assumption that both sy607 and sy275 phenotypes are caused by different reduction of function of the cog-1 gene. Filled boxes with red or blue outline indicate ectopic expression. (G) Altered pattern of gene expression in lin-11 mutants (26). (H) Altered pattern of gene expression in the egl-38 mutant (16) (Tables 1 and 2). egl-17 expression in vulF is observed in the cog-1(sy275); egl-38 double mutant, suggesting a redundant repression mechanism.
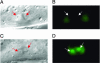
Fig. 2.
Regulation of egl-17 by cog-1. (A and B) Nomarski and epifluorescence images of wild-type mid-L4 animal carrying the egl-17::gfp transgene. Arrows point to vulE nuclei. vulE cells are not fluorescent. (C and D) cog-1(sy275) animals at the same stage carrying the egl-17::gfp transgene. vulE cells are fluorescent.
Fig. 3.
Link between cell fate patterning mechanisms and gene expression. In general, inductive signals regulate transcription factor networks to regulate gene expression. In the P7.p (but not P5.p) lineage, Wnt signals transduced by lin-17 and lin-18 control the pattern of cog-1 and lin-11 expression (25, 31). cog-1 and lin-11 in turn regulate egl-17 and cdh-3 expression (Table 3) (26). It has not been determined whether cog-1 and lin-11 regulate each other. In the P6.p lineage, an anchor cell signal and a let-60 Ras signal transduction pathway are required to establish the correct pattern of zmp-1 expression pattern (17). zmp-1 expression is also repressed in vulF by egl-38 PAX2/5/8 (Table 1). It is not known whether the patterning mechanism acts through egl-38. The expression pattern of egl-38 is also not known.
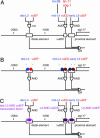
Fig. 4.
cis_-_regulatory elements of egl-17.(A) A map of the egl-17 5′ regulatory region. Boxes indicate enhancer elements defined by Cui and Han (34) and Kirouac and Sternberg (33). “AND” and “OR” logic gate symbols indicate sites and logic of information integration. Temporal (blue) and spatial (red) information is integrated as indicated by the logic circuit diagram to produce the complete egl-17 expression pattern. In one model (B), spatially and temporally regulated transcription factors each bind directly to the egl-17 cis-regulatory region. The integration of information takes place on enhancer elements. In the alternative model (C), spatial and temporal cues are integrated at the transcription factor level. These transcription factors (purple) with both spatially and temporally restricted activity regulate each enhancer element.
Similar articles
- The tailless ortholog nhr-67 regulates patterning of gene expression and morphogenesis in the C. elegans vulva.
Fernandes JS, Sternberg PW. Fernandes JS, et al. PLoS Genet. 2007 Apr 27;3(4):e69. doi: 10.1371/journal.pgen.0030069. Epub 2007 Mar 19. PLoS Genet. 2007. PMID: 17465684 Free PMC article. - Cell fates and fusion in the C. elegans vulval primordium are regulated by the EGL-18 and ELT-6 GATA factors -- apparent direct targets of the LIN-39 Hox protein.
Koh K, Peyrot SM, Wood CG, Wagmaister JA, Maduro MF, Eisenmann DM, Rothman JH. Koh K, et al. Development. 2002 Nov;129(22):5171-80. doi: 10.1242/dev.129.22.5171. Development. 2002. PMID: 12399309 - lin-1 has both positive and negative functions in specifying multiple cell fates induced by Ras/MAP kinase signaling in C. elegans.
Tiensuu T, Larsen MK, Vernersson E, Tuck S. Tiensuu T, et al. Dev Biol. 2005 Oct 1;286(1):338-51. doi: 10.1016/j.ydbio.2005.08.007. Dev Biol. 2005. PMID: 16140291 - The SynMuv genes of Caenorhabditis elegans in vulval development and beyond.
Fay DS, Yochem J. Fay DS, et al. Dev Biol. 2007 Jun 1;306(1):1-9. doi: 10.1016/j.ydbio.2007.03.016. Epub 2007 Mar 20. Dev Biol. 2007. PMID: 17434473 Free PMC article. Review. - LET-23-mediated signal transduction during Caenorhabditis elegans development.
Sternberg PW, Lesa G, Lee J, Katz WS, Yoon C, Clandinin TR, Huang LS, Chamberlin HM, Jongeward G. Sternberg PW, et al. Mol Reprod Dev. 1995 Dec;42(4):523-8. doi: 10.1002/mrd.1080420422. Mol Reprod Dev. 1995. PMID: 8607985 Review.
Cited by
- Identifying the Caenorhabditis elegans vulval transcriptome.
Zhang Q, Hrach H, Mangone M, Reiner DJ. Zhang Q, et al. G3 (Bethesda). 2022 May 30;12(6):jkac091. doi: 10.1093/g3journal/jkac091. G3 (Bethesda). 2022. PMID: 35551383 Free PMC article. - A novel candidate cis-regulatory motif pair in the promoters of germline and oogenesis genes in C. elegans.
Linhart C, Halperin Y, Darom A, Kidron S, Broday L, Shamir R. Linhart C, et al. Genome Res. 2012 Jan;22(1):76-83. doi: 10.1101/gr.115626.110. Epub 2011 Sep 19. Genome Res. 2012. PMID: 21930893 Free PMC article. - Rapid sequence evolution of transcription factors controlling neuron differentiation in Caenorhabditis.
Jovelin R. Jovelin R. Mol Biol Evol. 2009 Oct;26(10):2373-86. doi: 10.1093/molbev/msp142. Epub 2009 Jul 9. Mol Biol Evol. 2009. PMID: 19589887 Free PMC article. - Temporal ChIP-on-chip reveals Biniou as a universal regulator of the visceral muscle transcriptional network.
Jakobsen JS, Braun M, Astorga J, Gustafson EH, Sandmann T, Karzynski M, Carlsson P, Furlong EE. Jakobsen JS, et al. Genes Dev. 2007 Oct 1;21(19):2448-60. doi: 10.1101/gad.437607. Genes Dev. 2007. PMID: 17908931 Free PMC article. - The transcription factor HLH-2/E/Daughterless regulates anchor cell invasion across basement membrane in C. elegans.
Schindler AJ, Sherwood DR. Schindler AJ, et al. Dev Biol. 2011 Sep 15;357(2):380-91. doi: 10.1016/j.ydbio.2011.07.012. Epub 2011 Jul 18. Dev Biol. 2011. PMID: 21784067 Free PMC article.
References
- Wood, W. B. (1988) The Nematode Caenorhabditis elegans (Cold Spring Harbor Lab. Press, Plainview, NY).
- Kimble, J. (1981) Dev. Biol. 87, 286–300. - PubMed
- Sternberg, P. W. & Horvitz, H. R. (1986) Cell 44, 761–772. - PubMed
- Sternberg, P. W. (1988) Nature 335, 551–554. - PubMed
- Yochem, J., Weston, K. & Greenwald, I. (1988) Nature 335, 547–550. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources