Analysis of genes induced by Sendai virus infection of mutant cell lines reveals essential roles of interferon regulatory factor 3, NF-kappaB, and interferon but not toll-like receptor 3 - PubMed (original) (raw)
Analysis of genes induced by Sendai virus infection of mutant cell lines reveals essential roles of interferon regulatory factor 3, NF-kappaB, and interferon but not toll-like receptor 3
Christopher P Elco et al. J Virol. 2005 Apr.
Abstract
Sendai virus (SeV) infection causes the transcriptional induction of many cellular genes that are also induced by interferon (IFN) or double-stranded RNA (dsRNA). We took advantage of various mutant cell lines to investigate the putative roles of the components of the IFN and dsRNA signaling pathways in the induction of those genes by SeV. Profiling the patterns of gene expression in SeV-infected cells demonstrated that Toll-like receptor 3, although essential for gene induction by dsRNA, was dispensable for gene induction by SeV. In contrast, Jak1, which mediates IFN signaling, was required for the induction of a small subset of genes by SeV. NF-kappaB and interferon regulatory factor 3 (IRF-3), the two major transcription factors activated by virus infection, were essential for the induction of two sets of genes by SeV. As expected, some of the IRF-3-dependent genes, such as ISG56, were more strongly induced by SeV in IRF-3-overexpressing cells. Surprisingly, in those cells, a number of NF-kappaB-dependent genes, such as the A20 gene, were induced poorly. Using a series of cell lines expressing increasing levels of IRF-3, we demonstrated that the degree of induction of A20 mRNA, upon SeV infection, was inversely proportional to the cellular level of IRF-3, whereas that of ISG56 mRNA was directly proportional. Thus, IRF-3 can suppress the expression of NF-kappaB-dependent genes in SeV-infected cells.
Figures
FIG. 1.
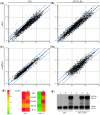
Effect of TLR3 on the regulation of cellular genes by SeV and dsRNA. Shown are global changes in mRNA levels in 293 cells (A and C) or 293 cells expressing TLR3 (B and D) 6 h after SeV infection at 1 hemagglutinating unit/4.0 × 103 cells (A and B) or dsRNA treatment at 100 μg/ml (C and D) as determined by cDNA microarray experiment. Each point on the scatter plots represents the expression of an individual mRNA message, as determined by units of fluorescent intensity, in untreated cells (x axis) plotted against its expression 6 h after SeV infection or dsRNA treatment (y axis). The central diagonal line (black) represents equal expression in treated and untreated samples, while twofold differences in expression are indicated by the two flanking blue lines. (E) Regulation of specific genes by dsRNA and SeV in 293 (−) and 293/TLR3 (+) cells. The tiles show the increase (_n_-fold) in mRNA expression for specific genes in SeV- or dsRNA-treated cells relative to untreated cells as a function of color. Green, expression was unchanged; yellow to red, expression was induced to increasing degrees. (F) RPA of ISG56 induction in untreated (lanes 1 and 4), dsRNA-treated (lanes 2 and 5), or SeV-infected (lanes 3 and 6) 293 cells (lanes 1 to 3) or 293/TLR3 cells (lanes 4 to 6).
FIG. 2.
Differential induction of genes by SeV and dsRNA. Select genes were differentially regulated by SeV and dsRNA in 293 cells expressing (+) or not expressing (−) TLR3. Colors represent induction (_n_-fold) by SeV (left column) or dsRNA (right column) as described in the legend to Fig. 1.
FIG. 3.
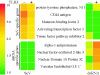
IFN-dependent regulation of SeV-induced genes. Select SeV-regulated genes display differential induction patterns in wt (2fTGH) and Jak1−/− (U4C) cells. (A) Microarray data showing average increase (_n_-fold) in mRNA expression 6 h after SeV infection in 2fTGH and U4C cells. Genes are grouped based on their independence of, dependence on, or impairment by Jak1 for induction. (B) RPA (A20) and Northern (OAS, p69) analysis of mRNA expression in 2fTGH and U4C cells, before or 6 h after SeV infection.
FIG. 4.
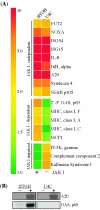
Requirement for NF-κB for gene induction by SeV. Genes were regulated by SeV in wt (2fTGH) and NF-κB null (2F-SR) cells. (A) Average induction (_n_-fold) of SeV-regulated genes, grouped by dependence on NF-κB, as determined by microarray. (B) Quantitative analysis of NOXA, ISG56, and A20 mRNA induction by RPA.
FIG. 5.
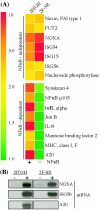
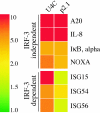
IRF-3-dependent gene induction. Shown are examples of genes regulated by SeV in U4C cells and P2.1 cells in which IRF-3 expression is impaired. Genes are grouped by their dependence on IRF-3 for induction.
FIG. 6.
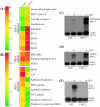
Regulation of SeV-induced genes by IRF-3 and altered expression of SeV-regulated genes in U4C cells and U4C.2 cells, which express high levels of IRF-3. (A and B) Microarray data for the induction (_n_-fold) of select SeV-regulated genes in U4C and U4C.2 cells. Genes are grouped as follows: IRF-3 enhanced, genes with augmented expression in IRF-3-overexpressing cells (i.e., induced more strongly in U4C.2 cells); IRF-3 neutral, genes unaffected by cellular IRF-3 levels (induced at equivalent levels in both cell lines); IRF-3 repressed, SeV-induced genes negatively regulated by IRF-3 (induced in U4C but not U4C.2 cells). Also shown are quantitative RPA analyses of follistatin (C), NOXA (D), and A20 (E) mRNA induction in U4C and U4C.2 cells before or after SeV infection.
FIG. 7.
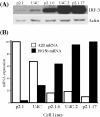
Modulation of A20 mRNA and ISG56 mRNA expression by cellular levels of IRF-3. Cell lines derived from U4C and P2.1 cells and expressing different levels of IRF-3 protein were infected with SeV and analyzed for the expression of A20 mRNA and ISG56 mRNA. (A) Western blot showing the relative levels of IRF-3 expression in the different cell lines relative to actin. (B) Percent maximum induction (_n_-fold) of A20 mRNA (white bars) and ISG56 mRNA (black bars) in cells 6 h after infection, normalized to actin mRNA expression as determined by RPA.
FIG. 8.
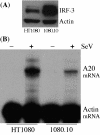
Negative regulation of SeV-induced A20 mRNA expression by IRF-3 in wt cells. (A) Western analysis of IRF-3 expression in HT1080 and 1080.10 cells. (B) RPA comparing A20 mRNA induction in untreated (−) and SeV-infected (+) HT1080 and 1080.10 cells.
Similar articles
- Convergence of the NF-kappaB and interferon signaling pathways in the regulation of antiviral defense and apoptosis.
Hiscott J, Grandvaux N, Sharma S, Tenoever BR, Servant MJ, Lin R. Hiscott J, et al. Ann N Y Acad Sci. 2003 Dec;1010:237-48. doi: 10.1196/annals.1299.042. Ann N Y Acad Sci. 2003. PMID: 15033728 Review. - Regulation of IFN regulatory factor-7 and IFN-alpha production by enveloped virus and lipopolysaccharide in human plasmacytoid dendritic cells.
Dai J, Megjugorac NJ, Amrute SB, Fitzgerald-Bocarsly P. Dai J, et al. J Immunol. 2004 Aug 1;173(3):1535-48. doi: 10.4049/jimmunol.173.3.1535. J Immunol. 2004. PMID: 15265881 - A20 is a potent inhibitor of TLR3- and Sendai virus-induced activation of NF-kappaB and ISRE and IFN-beta promoter.
Wang YY, Li L, Han KJ, Zhai Z, Shu HB. Wang YY, et al. FEBS Lett. 2004 Oct 8;576(1-2):86-90. doi: 10.1016/j.febslet.2004.08.071. FEBS Lett. 2004. PMID: 15474016 - Ectopic expression of toll-like receptor-3 (TLR-3) overcomes the double-stranded RNA (dsRNA) signaling defects of P2.1 cells.
Sun Y, Leaman DW. Sun Y, et al. J Interferon Cytokine Res. 2004 Jun;24(6):350-61. doi: 10.1089/107999004323142213. J Interferon Cytokine Res. 2004. PMID: 15212709 - Transcriptional signaling by double-stranded RNA: role of TLR3.
Sen GC, Sarkar SN. Sen GC, et al. Cytokine Growth Factor Rev. 2005 Feb;16(1):1-14. doi: 10.1016/j.cytogfr.2005.01.006. Cytokine Growth Factor Rev. 2005. PMID: 15733829 Review.
Cited by
- Interferon regulatory factor 3 inhibits astrocyte inflammatory gene expression through suppression of the proinflammatory miR-155 and miR-155*.
Tarassishin L, Loudig O, Bauman A, Shafit-Zagardo B, Suh HS, Lee SC. Tarassishin L, et al. Glia. 2011 Dec;59(12):1911-22. doi: 10.1002/glia.21233. Glia. 2011. PMID: 22170100 Free PMC article. - Viral induction of the zinc finger antiviral protein is IRF3-dependent but NF-kappaB-independent.
Wang N, Dong Q, Li J, Jangra RK, Fan M, Brasier AR, Lemon SM, Pfeffer LM, Li K. Wang N, et al. J Biol Chem. 2010 Feb 26;285(9):6080-90. doi: 10.1074/jbc.M109.054486. Epub 2010 Jan 4. J Biol Chem. 2010. PMID: 20048147 Free PMC article. - TLR3 and TLR4 are innate antiviral immune receptors in human microglia: role of IRF3 in modulating antiviral and inflammatory response in the CNS.
Suh HS, Zhao ML, Choi N, Belbin TJ, Brosnan CF, Lee SC. Suh HS, et al. Virology. 2009 Sep 30;392(2):246-59. doi: 10.1016/j.virol.2009.07.001. Epub 2009 Jul 30. Virology. 2009. PMID: 19646728 Free PMC article. - The ISG56/IFIT1 gene family.
Fensterl V, Sen GC. Fensterl V, et al. J Interferon Cytokine Res. 2011 Jan;31(1):71-8. doi: 10.1089/jir.2010.0101. Epub 2010 Oct 15. J Interferon Cytokine Res. 2011. PMID: 20950130 Free PMC article. Review. - IRF-3 activation by Sendai virus infection is required for cellular apoptosis and avoidance of persistence.
Peters K, Chattopadhyay S, Sen GC. Peters K, et al. J Virol. 2008 Apr;82(7):3500-8. doi: 10.1128/JVI.02536-07. Epub 2008 Jan 23. J Virol. 2008. PMID: 18216110 Free PMC article.
References
- Alexopoulou, L., A. C. Holt, R. Medzhitov, and R. A. Flavell. 2001. Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature 413:732-738. - PubMed
- Bandyopadhyay, S. K., G. T. Leonard, Jr., T. Bandyopadhyay, G. R. Stark, and G. C. Sen. 1995. Transcriptional induction by double-stranded RNA is mediated by interferon-stimulated response elements without activation of interferon-stimulated gene factor 3. J. Biol. Chem. 270:19624-19629. - PubMed
- Boone, D. L., E. E. Turer, E. G. Lee, R. C. Ahmad, M. T. Wheeler, C. Tsui, P. Hurley, M. Chien, S. Chai, O. Hitotsumatsu, E. McNally, C. Pickart, and A. Ma. 2004. The ubiquitin-modifying enzyme A20 is required for termination of Toll-like receptor responses. Nat. Immunol. 5:1052-1060. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 AI034039/AI/NIAID NIH HHS/United States
- GM07250/GM/NIGMS NIH HHS/United States
- CA68782/CA/NCI NIH HHS/United States
- AI34039/AI/NIAID NIH HHS/United States
- CA62220/CA/NCI NIH HHS/United States
- P01 CA062220/CA/NCI NIH HHS/United States
- T32 GM007250/GM/NIGMS NIH HHS/United States
- R01 CA068782/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous