Inhibition of RIG-I-dependent signaling to the interferon pathway during hepatitis C virus expression and restoration of signaling by IKKepsilon - PubMed (original) (raw)
Inhibition of RIG-I-dependent signaling to the interferon pathway during hepatitis C virus expression and restoration of signaling by IKKepsilon
Adrien Breiman et al. J Virol. 2005 Apr.
Abstract
Interferon (IFN) is one important effector of the innate immune response, induced by different viral or bacterial components through Toll-like receptor (TLR)-dependent and -independent mechanisms. As part of its pathogenic strategy, hepatitis C virus (HCV) interferes with the innate immune response and induction of IFN-beta via the HCV NS3/4A protease activity which inhibits phosphorylation of IRF-3, a key transcriptional regulator of the IFN response. In the present study, we demonstrate that inhibition by the protease occurs upstream of the noncanonical IKK-related kinases IKKepsilon and TBK-1, which phosphorylate IRF-3, through partial inhibition of the TLR adapter protein TRIF/TICAM1-dependent pathway. Use of TRIF(-/-) mouse embryo fibroblasts however revealed the presence of a TRIF-independent pathway involved in IFN induction that was also inhibited by NS3/4A. Importantly, we show that NS3/4A can strongly inhibit the ability of the recently described RIG-I protein to activate IFN, suggesting that RIG-I is a key factor in the TRIF-independent, NS3/4A-sensitive pathway. Expression of IFN signaling components including IKKepsilon, TBK-1, TRIF, and wild type or constitutively active forms of RIG-I in the HCV replicon cells resulted in IFN-beta promoter transactivation, with IKKepsilon displaying the highest efficiency. Subsequently, overexpression of IKKepsilon resulted in 80% inhibition of both the positive and negative replicative strands of the HCV replicon. The partial restoration of the capacity of the host cell to transcribe IFN-beta indicates that IKKepsilon expression is able to bypass the HCV-mediated inhibition and restore the innate antiviral response.
Figures
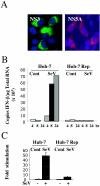
FIG. 1.
IFN production is inhibited in an HCV replicon-expressing cell. (A) Huh-7 Rep cells were plated at 20,000 cells/well in Labtek chambers. After 24 h of incubation, the cells were fixed with 3.7% paraformaldehyde, permeabilized with Triton X-100, and analyzed by immunofluorescence for the presence of HCV proteins with a monoclonal anti-NS3 (a gift from D.Moradpour) and fluorescein isothiocyanate-conjugated anti-mouse immunoglobulin antibodies or monoclonal anti-NS5A (Biodesign) and Texas Red-conjugated anti-mouse immunoglobulin antibodies. (B) Huh-7 and Huh-7 Rep cells were plated in six-well plates at 800,000 cells/well and 106 cells/well, respectively. At 24 h, the cells were infected with Sendaï virus (SeV) at 40 hemagglutination units (HAU)/well. At 4, 8, and 24 h after infection, infected and control (Cont) cells were washed twice with phosphate-buffered saline, RNA was extracted with RNABle, and the samples were processed for real-time RT-PCR analysis as described in Materials and Methods. For each sample, reverse transcription was conducted in the simultaneous presence of the 3′ primers for GAPDH and IFN-β to minimize errors. All values were normalized with GAPDH. (C) Huh-7 and Huh-7 Rep cells were transfected with 1 μg of IFN-β-pGL3 and 0.4 μg of a plasmid expressing Rous sarcoma virus β-galactosidase for normalization. After 8 h, cells were further infected with Sendaï virus at 40 hemagglutination units (HAU)/ml. After 24 h, the cell extracts were analyzed for luciferase activity in duplicate, andthe results are expressed as stimulation (n = fold) of luciferase activity after β-galactosidase normalization.
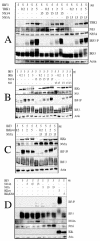
FIG. 2.
HCV NS3/4A protease does not affect IKKɛ/TBK-1-mediated IRF-3 phosphorylation. HEK 293T cells were seeded at 3.5 × 106 cells/100-mm plate and transfected after 24 h with Lipofectamine 2000 with 5 μg of plasmid expressing IRF-3 in the presence of (A) different concentrations of a plasmid expressing TBK-1, in either the absence or the presence of 15 μg of plasmids expressing NS3/4A or NS5A; (B) different concentrations of a plasmid expressing wild-type IKKɛ, either in the absence or in the presence of 15 μg of plasmids expressing NS3/4Aor NS3 alone; (C) different concentrations of a plasmid expressing wild-type IKK, in the presence of 15 μg of plasmid expressing NS5A; or (D) different amounts of plasmids expressing NS3/4A, NS5A, wild-type IKKɛ, or IKKɛ K38A. Cell extracts were prepared 24 h after transfection, and equivalent amounts of protein extract (45 μg) were used for immunoblot analysis.
FIG. 3.
NS3/4A affects IFN signaling via TRIF-dependent and TRIF-independent pathways. (A) HEK 293T cells were cotransfected with the IFN-β-pGL3 reporter construct and the PRLTK internal control in the presence or absence of NS3/4A and TRIF expression plasmids as described in Materials and Methods. (B) HEK 293 T cells were cotransfected with an empty vector (pcDNA3/Amp) or plasmid expressing TRIF (1.2 μg) alone or in the presence of 2,5 μg of plasmid expressing either NS3/4A or NS5A; 24 h after the transfection, RNA was extracted with RNABle, and the samples were processed for real-time RT-PCR analysis as described in Materials and Methods and for Fig. 1B. (C) Wild-type MEFs (white bars) and TRIF−/− MEFs (dark bars) were transfected with the IFN-β-pGL3 reporter construct and the PRLTK internal control in the presence or absence of NS3/4A and NS5A expression plasmids. Cells were either infected with Sendai virus or left untreated. Analysis of luciferase activity was measured 24 h after transfection and normalized with the Renilla luciferase activities. An immunoblot shows the presence of TRIF in the wild-type MEFs (+/+) and its absence in the TRIF−/− MEFs. (D) HEK 293 T cells were cotransfected with the IFN-β-pGL3 reporter construct (100 ng) and the PRLTK internal control in the presence or absence of 300 ng of ΔRIG-1, 300 ng of a dominant negative form of IKKɛ deprived of its catalytic domain (IKKɛ ΔC), and 300 ng of NS3/4A or NS5A expression plasmid. When indicated, the cells were infected with 40 hemagglutination units (HAU) of Sendai virus/106 cells for 16 h.
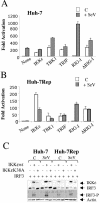
FIG. 4.
Relative efficiency of different components of IFN signaling to restore activation of the IFN-β promoter in the HCV replicon cells. (A and B) Huh-7 (A) and Huh-7 Rep (B) cells were transfected with 1 μg of IFN-β-pGL3, 1 μg of Rous sarcoma virus β-galactosidase as such or in the presence of 0.5 μg of IKKɛ, TBK-1, TRIF, RIG-I, or ΔRIG-I. The stimulation of luciferase activity was analyzed without (empty bars) or after infection with Sendaï virus at 40 hemagglutination units (HAU)/ml (grey bars). Analysis of luciferase activity was performed as described above. (C) Huh-7 cells and Huh-7 Rep cells were transfected with 5 μg of plasmids expressing IRF-3, wild-type IKKɛ, or IKKɛ K38A, as indicated. At 24 h after transfection, the cells were infected with 80 hemagglutination units (HAU)/ml of Sendai virus for 8 h. Cell extracts were prepared and equivalent amounts of protein extract (30 μg) were used for immunoblot analysis.
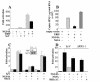
FIG. 5.
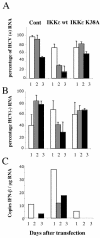
Overexpression of wild-type IKKɛ inhibits replication of an HCV replicon and partially restores IFN induction. Huh-7 Rep cells were plated in 100-mm plates at 2 × 106 cells/dish and transfected 24 h after seeding with 10 μg of an empty vector (Cont) or vectors expressing wild-type IKKɛ or IKKɛ K38A. At 24 h (1), 48 h (2), and 72 h (3) after transfection, RNA was extracted with RNABle, and the samples were processed for real-time RT-PCR analysis of HCV positive- (A) or negative-strand (B) RNA and of IFN-β RNA (C) as described in Materials and Methods. For each sample, reverse transcription was conducted in the simultaneous presence of the 3′ primers for GAPDH and HCV positive strand (A), GAPDH and HCV negative strand (B), and GAPDH and IFN-β (C) to minimize errors. The effect of IKKɛ on HCV RNA expression was analyzed in two independent experiments, and the results are expressed as the mean percent HCV RNA expression.
Similar articles
- Induction of IRF-3 and IRF-7 phosphorylation following activation of the RIG-I pathway.
Paz S, Sun Q, Nakhaei P, Romieu-Mourez R, Goubau D, Julkunen I, Lin R, Hiscott J. Paz S, et al. Cell Mol Biol (Noisy-le-grand). 2006 May 15;52(1):17-28. Cell Mol Biol (Noisy-le-grand). 2006. PMID: 16914100 - Hepatitis A virus suppresses RIG-I-mediated IRF-3 activation to block induction of beta interferon.
Fensterl V, Grotheer D, Berk I, Schlemminger S, Vallbracht A, Dotzauer A. Fensterl V, et al. J Virol. 2005 Sep;79(17):10968-77. doi: 10.1128/JVI.79.17.10968-10977.2005. J Virol. 2005. PMID: 16103148 Free PMC article. - Limited suppression of the interferon-beta production by hepatitis C virus serine protease in cultured human hepatocytes.
Dansako H, Ikeda M, Kato N. Dansako H, et al. FEBS J. 2007 Aug;274(16):4161-76. doi: 10.1111/j.1742-4658.2007.05942.x. Epub 2007 Jul 25. FEBS J. 2007. PMID: 17651439 - The interferon inducing pathways and the hepatitis C virus.
Meurs EF, Breiman A. Meurs EF, et al. World J Gastroenterol. 2007 May 7;13(17):2446-54. doi: 10.3748/wjg.v13.i17.2446. World J Gastroenterol. 2007. PMID: 17552028 Free PMC article. Review. - The host type I interferon response to viral and bacterial infections.
Perry AK, Chen G, Zheng D, Tang H, Cheng G. Perry AK, et al. Cell Res. 2005 Jun;15(6):407-22. doi: 10.1038/sj.cr.7290309. Cell Res. 2005. PMID: 15987599 Review.
Cited by
- Modulation of Kinase Activities In Vitro by Hepatitis C Virus Protease NS3/NS4A Mediated-Cleavage of Key Immune Modulator Kinases.
Abdullah MAF, McWhirter SM, Suo Z. Abdullah MAF, et al. Cells. 2023 Jan 25;12(3):406. doi: 10.3390/cells12030406. Cells. 2023. PMID: 36766748 Free PMC article. - Effect of protease and helicase mutations on HCV NS3 activity.
Eisa ZM. Eisa ZM. Saudi J Biol Sci. 2011 Apr;18(2):195-200. doi: 10.1016/j.sjbs.2010.09.001. Epub 2010 Oct 14. Saudi J Biol Sci. 2011. PMID: 23961124 Free PMC article. - Encephalomyocarditis virus 2A protein is required for viral pathogenesis and inhibition of apoptosis.
Carocci M, Cordonnier N, Huet H, Romey A, Relmy A, Gorna K, Blaise-Boisseau S, Zientara S, Kassimi LB. Carocci M, et al. J Virol. 2011 Oct;85(20):10741-54. doi: 10.1128/JVI.00394-11. Epub 2011 Aug 17. J Virol. 2011. PMID: 21849462 Free PMC article. - Bovine Adenovirus-3 pVIII Suppresses Cap-Dependent mRNA Translation Possibly by Interfering with the Recruitment of DDX3 and Translation Initiation Factors to the mRNA Cap.
Ayalew LE, Patel AK, Gaba A, Islam A, Tikoo SK. Ayalew LE, et al. Front Microbiol. 2016 Dec 27;7:2119. doi: 10.3389/fmicb.2016.02119. eCollection 2016. Front Microbiol. 2016. PMID: 28082972 Free PMC article. - Human metapneumovirus glycoprotein G disrupts mitochondrial signaling in airway epithelial cells.
Bao X, Kolli D, Ren J, Liu T, Garofalo RP, Casola A. Bao X, et al. PLoS One. 2013 Apr 23;8(4):e62568. doi: 10.1371/journal.pone.0062568. Print 2013. PLoS One. 2013. PMID: 23626834 Free PMC article.
References
- Abbate, I., M. Romano, R. Longo, G. Cappiello, O. Lo Iacono, V. Di Marco, C. Paparella, A. Spano, and M. R. Capobianchi. 2003. Endogenous levels of mRNA for IFNs and IFN-related genes in hepatic biopsies of chronic HCV-infected and non-alcoholic steatohepatitis patients. J. Med. Virol. 70:581-587. - PubMed
- Alexopoulou, L., A. C. Holt, R. Medzhitov, and R. A. Flavell. 2001. Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature 413:732-738. - PubMed
- Bain, C., A. Fatmi, Z. Zoulim, J. P. Zarski, C. Trepo, and G. Inschauspe. 2001. Impaired allostimulatory function of dendritic cells in chronic hepatitis C infection. Gastroenterology 120:512-524. - PubMed
- Bartenschlager, B. 1999. The NS3/4A proteinase of the hepatitis C virus: unravelling structure and function of an unusual enzyme and a prime target for antiviral therapy. J. Viral Hepatitis 6:165-181. - PubMed
- Bartenschlager, R., and V. Lohmann. 2001. Novel cell cultures systems for the hepatitis C virus. Antiviral Res. 52:1-17. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous