Dual role of the lymphocytic choriomeningitis virus intergenic region in transcription termination and virus propagation - PubMed (original) (raw)
Dual role of the lymphocytic choriomeningitis virus intergenic region in transcription termination and virus propagation
Daniel D Pinschewer et al. J Virol. 2005 Apr.
Abstract
Each genome segment of the prototypic arenavirus lymphocytic choriomeningitis virus (LCMV), encodes two genes in ambisense orientation, separated by an intergenic region (IGR). The 3' ends of subgenomic viral mRNAs have been mapped to a stem-loop structure within the IGR, suggesting structure-dependent transcription termination. We have studied the role of the LCMV IGR by using a minigenome (MG) rescue system based on RNA analogues of the short genome segment. An ambisense MG coding for chloramphenicol acetyltransferase (CAT) and green fluorescent protein reporter genes instead of the nucleoprotein and glycoprotein open reading frames, respectively, served as a template for synthesis of full-length anti-MG (aMG) replicate and subgenomic size mRNA for reporter gene expression. An analogous MG without IGR was amplified by the virus polymerase with equal efficiency, but subgenomic mRNA was undetectable. Reporter gene expression from IGR-deficient aMG CAT-sense RNA of genomic length was approximately 5-fold less efficient than that from subgenomic CAT mRNA derived from an IGR-containing MG, but at least 100-fold more efficient than that from a T7 RNA polymerase transcript with the same sequence. Therefore, in the absence of IGR-mediated transcription termination, a fraction of full-length aMG RNA appears to behave as bona fide mRNA. Unexpectedly, MGs without IGR were dramatically impaired in their ability to passage reporter gene activity via infectious virus-like particles. These data suggest that the LCMV IGR serves individual functions in transcription termination for enhanced gene expression and in the virus assembly and/or budding, which are required for the efficient propagation of LCMV infectivity.
Figures
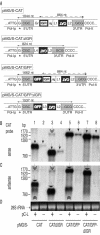
FIG. 1.
Effects of the IGR on MG RNA expression. (A) pMG/S-CAT (previously referred to as pMG-ARM/S) (25) contains the MG/S-CAT construct flanked by the murine Pol-I promoter (Pol-Ip) and Pol-I terminator (Pol-It). Transcription of pMG/S-CAT by the cellular Pol-I (primary transcription) generates MG/S-CAT RNA with a 5′ UTR containing a nontemplated G residue (*) (29) and a 3′ UTR containing the precise viral 3′ end (see also Fig. 5). Constructs pMG/S-CATΔIGR, pMG/S-CAT/GFP, and pMG/S-CAT/GFPΔIGR contain the indicated RNA analogues, flanked also by the murine Pol-Ip and Pol-It. Inverted writing indicates antisense polarity with respect to Pol-Ip. Gr and Nr, sequences derived from residual GP (90 nucleotides [nt]) and NP ORF (55 nt), respectively; L1 to L4, linker sequences for cloning purposes (6 to 13 nt in length). Lines with arrows at both ends indicate the lengths of primary Pol-I transcripts (solid lines) or ofsubgenomic mRNA transcripts formed by the LCMV RdRp (dashed lines; in approximation) (20), where applicable. (B to D) BHK-21 cells in six-well plates (80% confluent) were transfected with the indicated MG expression vectors (0.5 μg) together with pC-NP (0.8 μg), pC-GP (0.4 μg), and pC-Z (0.1 μg) by using Lipofectamine as described previously (6). pC-L (1 μg) was added to the transfection mix where indicated. Sixty hours later, total cellular RNA was prepared as described previously (6). Duplicate Northern blots were hybridized to CAT sense (B) and CAT antisense (C) riboprobes. Ethidium bromide staining of 28S rRNA showed comparable total RNA amounts loaded for each sample (D). Narrow solid arrows, MG polarity full-length RNA; wide solid arrows, anti-MG polarity full-length RNA; double-headed arrows, putative MG homodimers as previously described (14); triple-headed arrows, CAT mRNA of subgenomic length. Samples originate from the same experiment as that shown in Fig. 2A and B and 4A and B.
FIG. 2.
Impact of the IGR on MG reporter gene expression. BHK-21 cells in six-well plates (80% confluent) were transfected with the indicated MG expression vectors (0.5 μg) together with pC-NP (0.8 μg), pC-GP (0.4 μg), and pC-Z (0.1 μg) by using Lipofectamine as described previously (6). pC-L (1 μg) was added to the transfection mix where indicated. The chart in panel A is representative of the correspondingly numbered samples in panel B. Sixty hours later, the cultures were monitored for GFP expression by fluorescence microscopy (panel B, samples 5 to 8 only) and cell extracts were processed for the CAT assay (A) as described previously (6). O, origin of sample application; NAc, nonacetylated chloramphenicol; MAc, monoacetylated chloramphenicol. The samples originate from the same experiment as that shown in Fig. 1B to D and 4A and B.
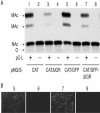
FIG. 3.
Inefficient CAT translation from T7-derived full-length anti-MG RNA. (A) Construct pT7-aMGhrz expresses an antisense transcript of MG/S-CAT/GFPΔIGR under control of the T7 RNA polymerase promoter (T7p) and followed downstream by a sequence-adapted hairpin ribozyme (hrz) to generate the precise viral 3′ end and the T7 RNA polymerase terminator (T7T). Construct pT7-aMGdrz contains the hepatitis delta ribozyme (drz) instead of the hairpin ribozyme in pT7-aMGhrz. The drz-based construct contains an extra 3′ C residue (*) for efficient self-cleavage of drz (24). Inverted writing indicates antisense polarity with respect to T7p. L2 and L3, linker sequences for cloning purposes (6 to 13 nt in length); lines with arrows at both ends indicate the lengths of processed T7 transcripts. (B to E) BHK-21 cells in six-well plates (80% confluent) were transfected with the indicated amounts of pC-L, pC-NP (0.8 μg), pMG/S-CAT/GFPΔIGR (0.5 μg), pT7-MGhrz (0.5 μg), pT7-MGdrz (0.5 μg), andpC-T7 (1 μg) as indicated in the chart by using Lipofectamine as described previously (6). Sixty hours later, cell extracts were processed for the CAT assay (E) as previously described (6), and total cellular RNA from each sample was probed by Northern hybridization with a CAT antisense riboprobe (C and D) and exposed for 8 (C) or 40 (D) min, to detect and discriminate the individual bands in each lane. Ethidium bromide staining of 28S rRNA showed comparable total RNA amounts loaded for each sample (B). O, origin of sample application; NAc, nonacetylated chloramphenicol; MAc, monoacetylated chloramphenicol; *, unprocessed T7 transcript (lanes 4 and 5); **, T7 transcript upon self-processing of the respective ribozyme (lanes 4 and 5) or anti-MG RNA plus putative full-length antigenomic CAT mRNA-like transcript (lanes 2 and 3).
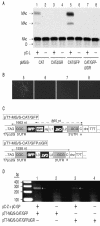
FIG. 4.
Effects of the IGR on VLP formation (A and B) BHK-21 cells in six-well plates (80% confluent) were transfected with MG expression vectors (0.5 μg) together with pC-NP (0.8 μg), pC-GP (0.4 μg) and pC-Z (0.1 μg) by using Lipofectamine as described previously (6). pC-L (1 μg) was added to the transfection mix where indicated. The chart in panel A is representative of the correspondingly numbered samples in panel B. O, origin of sample application; NAc, nonacetylated chloramphenicol; MAc, monoacetylated chloramphenicol. Sixty hours later, the cultures supernatants (400 μl [each] of the 2-ml total) were passaged onto fresh BHK-21 cell monolayers for 2 h prior to superinfection of the cultures with LCMV Armstrong at a multiplicity of infection of 2. Twenty-four h after passage, the cultures were monitored for GFP expression by fluorescence microscopy (panel B,samples 5 to 8 only). Seventy-two hours after passage, cell extracts prepared from the passage culture wells were processed for CAT assay (A). Samples originate from the same experiment as that shown in Fig. 1B to D and 2A and B. (C) Constructs pT7-MG/S-CAT/GFP and pT7-MG/S-CAT/GFPΔIGR express the respective MG under control of a modified T7 promoter (T7pΔ2G) (23), followed downstream by the hepatitis delta ribozyme (drz) to generate the precise viral 3′ end, and the T7 RNA polymerase terminator (T7T). An extra 3′ C residue (*) upstream of drz allows for efficient self-cleavage (24). Inverted writing indicates antisense polarity with respect to T7p. IGR, intergenic region; Nr, residual NP ORF-derived sequence (55 nt); L1 to L3, linker sequences for cloning purposes (6 to 13 nt in length). Lines with arrows at both ends indicate the lengths of primary Pol-I transcripts (solid lines) or of subgenomic mRNA transcripts formed by the LCMV RdRp (dashed lines; in approximation) (20) where applicable (compare Fig. 1C). (D) 293T cells in six-well plates (80% confluent) were transfected with pC-L (1 μg), pC-NP (0.8 μg), and pC-T7 (1 μg) by using Lipofectamine as described previously (6). pT7-MG/S-CAT/GFP (0.5 μg), pT7-MG/S-CAT/GFPΔIGR (0.5 μg), pC-GP (0.4 μg), and pC-Z (0.1 μg) were added to the transfection mixture as indicated in the chart. Seventy hours later, VLPs in the SN were purified by ultracentrifugation, and MG RNA was extracted and detected by RT-PCR as previously described (23). PCR products were resolved by gel electrophoresis and were detected by ethidium bromide staining. The arrows indicate the expected 280-bp CAT PCR product (CAT).
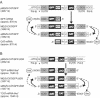
FIG. 5.
Schematic for postulated MG transcription and replication on IGR-competent and -deficient MG templates. (A) The MG template is synthesized intracellularly via nuclear Pol-I-mediated primary transcription (arrow with double lines) and exported to the cytoplasm via unknown mechanisms. Upon encapsidation by NP, distinct transcriptase (arrows with dashed lines) and replicase complexes (solid arrow) of the LCMV RdRp synthesize full-length replicate and subgenomic mRNA. Thereby, replication introduces a 5′ nontemplated G residue (29) that is lost in the subsequent replication step. Transcription initiation with a host-derived primer results in the introduction of a 5′ cap structure and of a variable number of nontemplated nucleotides (20). Transcriptase complexes terminate upon synthesis of the IGR sequence via structure-dependent termination (20, 33). (B) IGR-deficient MG templates are subject to identical processes, but transcriptase complexes proceed to the 5′ end of the MG or aMG template. Due to a 5′ cap structure, the so-produced molecules are efficiently translated, but they are unlikely to serve as templates for further RNA synthesis as their cap structure is supposed to interfere with encapsidation.
Similar articles
- NP and L proteins of lymphocytic choriomeningitis virus (LCMV) are sufficient for efficient transcription and replication of LCMV genomic RNA analogs.
Lee KJ, Novella IS, Teng MN, Oldstone MB, de La Torre JC. Lee KJ, et al. J Virol. 2000 Apr;74(8):3470-7. doi: 10.1128/jvi.74.8.3470-3477.2000. J Virol. 2000. PMID: 10729120 Free PMC article. - Role of the virus nucleoprotein in the regulation of lymphocytic choriomeningitis virus transcription and RNA replication.
Pinschewer DD, Perez M, de la Torre JC. Pinschewer DD, et al. J Virol. 2003 Mar;77(6):3882-7. doi: 10.1128/jvi.77.6.3882-3887.2003. J Virol. 2003. PMID: 12610166 Free PMC article. - Arenavirus Genome Rearrangement for the Development of Live Attenuated Vaccines.
Cheng BY, Ortiz-Riaño E, de la Torre JC, Martínez-Sobrido L. Cheng BY, et al. J Virol. 2015 Jul;89(14):7373-84. doi: 10.1128/JVI.00307-15. Epub 2015 May 13. J Virol. 2015. PMID: 25972555 Free PMC article. - Reverse genetics of arenaviruses.
Lee KJ, de la Torre JC. Lee KJ, et al. Curr Top Microbiol Immunol. 2002;262:175-93. doi: 10.1007/978-3-642-56029-3_8. Curr Top Microbiol Immunol. 2002. PMID: 11987806 Review. No abstract available. - Multifunctional noncoding regions in the mammarenavirus genome.
Iwasaki M. Iwasaki M. Virology. 2025 Apr;605:110464. doi: 10.1016/j.virol.2025.110464. Epub 2025 Feb 24. Virology. 2025. PMID: 40022944 Review.
Cited by
- S RNA Intergenic Deletions Drive Viral Interference during Arenavirus Infections.
Hackbart M, López CB. Hackbart M, et al. bioRxiv [Preprint]. 2023 Nov 1:2023.10.31.564889. doi: 10.1101/2023.10.31.564889. bioRxiv. 2023. PMID: 37961573 Free PMC article. Preprint. - The PI3K/Akt pathway contributes to arenavirus budding.
Urata S, Ngo N, de la Torre JC. Urata S, et al. J Virol. 2012 Apr;86(8):4578-85. doi: 10.1128/JVI.06604-11. Epub 2012 Feb 15. J Virol. 2012. PMID: 22345463 Free PMC article. - Human hemorrhagic Fever causing arenaviruses: molecular mechanisms contributing to virus virulence and disease pathogenesis.
Shao J, Liang Y, Ly H. Shao J, et al. Pathogens. 2015 May 21;4(2):283-306. doi: 10.3390/pathogens4020283. Pathogens. 2015. PMID: 26011826 Free PMC article. Review. - Single nucleoprotein residue modulates arenavirus replication complex formation.
Knopp KA, Ngo T, Gershon PD, Buchmeier MJ. Knopp KA, et al. mBio. 2015 Apr 28;6(3):e00524-15. doi: 10.1128/mBio.00524-15. mBio. 2015. PMID: 25922393 Free PMC article. - Distinct Molecular Mechanisms of Host Immune Response Modulation by Arenavirus NP and Z Proteins.
Stott RJ, Strecker T, Foster TL. Stott RJ, et al. Viruses. 2020 Jul 21;12(7):784. doi: 10.3390/v12070784. Viruses. 2020. PMID: 32708250 Free PMC article. Review.
References
- Berg, J. M. 1986. Potential metal-binding domains in nucleic acid binding proteins. Science 232:485-487. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous