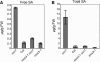A putative nucleoporin 96 Is required for both basal defense and constitutive resistance responses mediated by suppressor of npr1-1,constitutive 1 - PubMed (original) (raw)
A putative nucleoporin 96 Is required for both basal defense and constitutive resistance responses mediated by suppressor of npr1-1,constitutive 1
Yuelin Zhang et al. Plant Cell. 2005 Apr.
Abstract
The Arabidopsis thaliana suppressor of npr1-1, constitutive 1 (snc1) mutant contains a gain-of-function mutation in a Toll Interleukin1 receptor-nucleotide binding-Leu-rich repeat-type resistance gene (R-gene), which leads to constitutive activation of disease resistance response against pathogens. In a screen for suppressors of snc1, a recessive mutation, designated mos3 (for modifier of snc1,3), was found to suppress the constitutive pathogenesis-related gene expression and resistance to virulent Pseudomonas syringae maculicola ES4326 and Peronospora parasitica Noco2 in snc1. In addition, mos3 is also compromised in resistance mediated by Resistance to Peronospora parasitica 4 (RPP4), Resistance to Pseudomonas syringae pv maculicola (RPM1), and Resistance to Pseudomonas syringae 4 (RPS4). Single mutant mos3 plants exhibited enhanced disease susceptibility to P. s. pv maculicola ES4326, suggesting that MOS3 is required for basal resistance to pathogens as well. mos3-1 was identified by map-based cloning, and it encodes a protein with high sequence similarity to human nucleoporin 96. Localization of the MOS3-green fluorescent protein fusion to the nuclear envelope further indicates that MOS3 may encode a nucleoporin, suggesting that nuclear and cytoplasmic trafficking plays an important role in both R-gene-mediated and basal disease resistance.
Figures
Figure 1.
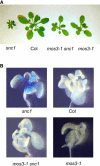
Phenotypic Analysis of the mos3-1 Mutant. (A) Morphology of snc1, the wild type (Col), mos3-1 snc1, and mos3-1. All plants were grown on soil and photographed when they were 4 weeks old. (B) Suppression of _snc1_-induced pBGL2-GUS reporter gene expression in mos3-1 snc1. Twenty-day-old seedlings grown on MS plates were stained for GUS activity as previously described (Bowling et al., 1994).
Figure 2.
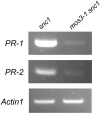
PR Gene Expression in mos3-1 snc1. RNAs were prepared from 20-d-old plants grown on MS media and reverse transcribed to obtain total cDNA. The cDNA samples were normalized by real-time PCR using an Actin1 probe. PR-1, PR-2, and Actin1 were amplified by 35 cycles of PCR using equal amounts of total cDNA, and the products were analyzed by agarose gel electrophoresis and ethidium bromide staining. The experiment was repeated once with the same results.
Figure 3.
SA Levels in snc1, Wild-Type (Col), mos3-1 snc1, and mos3-1 Plants. Leaves of 4-week-old soil-grown plants were collected, and SA was extracted and analyzed with high-pressure liquid chromatography using a previously described procedure (Li et al., 1999). (A), Free SA; (B), total SA levels in the leaves. The values presented are averages of four replicates ± standard deviations. The experiment was repeated once with similar results. FW, fresh weight.
Figure 4.
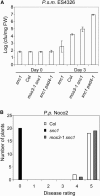
Growth of P.s.m. ES426 and P.p. Noco2 in snc1, Wild-Type (Col), snc1 pad4-1, and mos3-1 snc1 Plants. (A) Growth of P.s.m. ES4326. The leaves of 4-week-old soil-grown plants were infiltrated with a suspension of the bacteria at OD600 = 0.0001. Leaf discs within the inoculated areas were taken 3 d after infection. Four replicates were taken for each treatment. The experiment was repeated once with similar results. cfu, colony-forming units. (B) Growth of P.p. Noco2. Two-week-old seedlings were sprayed with Noco2 spores at a conidiospore suspension concentration of 3 × 104 spores per milliliter of water. The experiment was repeated twice with similar results. The infection was rated as follows on 20 plants 7 d after infection by counting the number of conidiophores per infected leaf: 0, no conidiophores on the plants; 1, no more than five conidiophores per infected leaf; 2, 6 to 20 conidiophores on a few of the infected leaves; 3, 6 to 20 conidiophores on most of the infected leaves; 4, five or more conidiophores on all infected leaves; 5, 20 or more conidiophores on all infected leaves.
Figure 5.
Growth of P.s.m. ES4326, P.p. Emoy2, P. syringae pv tomato DC3000 with AvrRps4, and P.s.m. ES4326 with AvrB on mos3-1. (A) Growth of P.s.m. ES4326 in mos3-1 and mos3-2 plants. The leaves of 4-week-old soil-grown plants were infiltrated with a suspension of the bacteria (OD600 = 0.0001). Leaf discs within the inoculated areas were taken 3 d after infection. Four replicates were taken for each treatment. cfu, colony-forming units; FW, fresh weight. (B) Growth of P.p. Emoy2 on mos3-1 plants. Two-week-old seedlings were sprayed with Emoy2 spores at a conidiospore suspension concentration of 5 × 104 spores per milliliter of water. The infection was rated as described in Figure 4 on 20 plants 7 d after infection by counting the number of conidiophores per infected leaf. (C) and (D) Growth of P. syringae pv tomato DC3000 avrRps4 (C) or P.s.m. ES4326 avrB (D) in mos3-1 plants. The leaves of 4-week-old soil-grown plants were infiltrated with a suspension of the bacteria (OD600 = 0.001). Leaf discs within the inoculated areas were taken 3 d after infection. Four replicates were taken for each treatment. All experiments were repeated at least twice with similar results.
Figure 6.
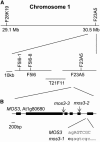
Map-Based Cloning of mos3-1 (A) Map of the mos3-1 locus on chromosome 1. Positions of the markers used for the mapping are indicated. Six recombinants pointed the mos3-1 mutation to the south of marker F5I6-1, and four recombinants pointed the mutation to the south of marker F5I6-8. (B) Exon/intron structure of MOS3. The coding regions are indicated with boxes. The position of the mos3-1 mutation is indicated, and its effects on the splicing of MOS3 are detailed below the wild-type sequence. Lowercase letters mark intron sequences, and uppercase letters indicate exon sequences. The arrows indicate the positions of T-DNA insertions in mos3-2 and mos3-3.
Figure 7.
Amino Acid Sequence Alignments of MOS3 (AtNup96), Human Nup96 (hNup96), and Nup145 of S. cerevisiae (ScNup145). Identical amino acids are shaded in black, and similar amino acids are shaded in gray. Alignment was performed using ClustalW (
http://www.ebi.ac.uk/clustalw/
). Boxshade was produced by BOXSHADE 3.21 (
http://www.ch.embnet.org/software/BOX\_form.html
). AtNup96, accession number NP_178183; hNup96, accession number NP_057404, amino acids 864 to 1800; ScNup145, accession number CAA54057, amino acids 606 to 1317.
Figure 8.
Complementation of mos3-1 snc1 by the MOS3 cDNA Clone. Two-week-old T3 plants homozygous for the MOS3 cDNA transgene in mos3-1 snc1 background were sprayed with P.p. Noco2 spores (5 × 104 spores/mL). The infection was rated as described in Figure 4 on 20 plants 7 d after infection by counting the number of conidiophores per infected leaf. The experiment was repeated once with similar results.
Figure 9.
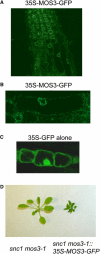
Subcellular Localization of MOS3. (A) Roots of 2-week-old Arabidopsis transgenic plants expressing the MOS3-GFP fusion protein were examined by confocal microscopy, and the picture was taken from the root tip region of a representative plant. (B) Magnified view of (A). Arabidopsis root cells expressing the MOS3-GFP fusion protein. (C) Two-week-old Arabidopsis root cells expressing GFP alone. (D) Complementation of mos3-1 with MOS3-GFP. Morphology of a representative T1 transgenic plant expressing MOS3-GFP protein in mos3-1 snc1 background.
Similar articles
- A gain-of-function mutation in a plant disease resistance gene leads to constitutive activation of downstream signal transduction pathways in suppressor of npr1-1, constitutive 1.
Zhang Y, Goritschnig S, Dong X, Li X. Zhang Y, et al. Plant Cell. 2003 Nov;15(11):2636-46. doi: 10.1105/tpc.015842. Epub 2003 Oct 23. Plant Cell. 2003. PMID: 14576290 Free PMC article. - An importin alpha homolog, MOS6, plays an important role in plant innate immunity.
Palma K, Zhang Y, Li X. Palma K, et al. Curr Biol. 2005 Jun 21;15(12):1129-35. doi: 10.1016/j.cub.2005.05.022. Curr Biol. 2005. PMID: 15964279 - Activation of an EDS1-mediated R-gene pathway in the snc1 mutant leads to constitutive, NPR1-independent pathogen resistance.
Li X, Clarke JD, Zhang Y, Dong X. Li X, et al. Mol Plant Microbe Interact. 2001 Oct;14(10):1131-9. doi: 10.1094/MPMI.2001.14.10.1131. Mol Plant Microbe Interact. 2001. PMID: 11605952 - Characterization of a novel, defense-related Arabidopsis mutant, cir1, isolated by luciferase imaging.
Murray SL, Thomson C, Chini A, Read ND, Loake GJ. Murray SL, et al. Mol Plant Microbe Interact. 2002 Jun;15(6):557-66. doi: 10.1094/MPMI.2002.15.6.557. Mol Plant Microbe Interact. 2002. PMID: 12059104 - Putative members of the Arabidopsis Nup107-160 nuclear pore sub-complex contribute to pathogen defense.
Wiermer M, Cheng YT, Imkampe J, Li M, Wang D, Lipka V, Li X. Wiermer M, et al. Plant J. 2012 Jun;70(5):796-808. doi: 10.1111/j.1365-313X.2012.04928.x. Epub 2012 Mar 6. Plant J. 2012. PMID: 22288649
Cited by
- Arabidopsis TRANSCURVATA1 encodes NUP58, a component of the nucleopore central channel.
Ferrández-Ayela A, Alonso-Peral MM, Sánchez-García AB, Micol-Ponce R, Pérez-Pérez JM, Micol JL, Ponce MR. Ferrández-Ayela A, et al. PLoS One. 2013 Jun 28;8(6):e67661. doi: 10.1371/journal.pone.0067661. Print 2013. PLoS One. 2013. PMID: 23840761 Free PMC article. - Identification of a maize locus that modulates the hypersensitive defense response, using mutant-assisted gene identification and characterization.
Chintamanani S, Hulbert SH, Johal GS, Balint-Kurti PJ. Chintamanani S, et al. Genetics. 2010 Mar;184(3):813-25. doi: 10.1534/genetics.109.111880. Epub 2010 Feb 22. Genetics. 2010. PMID: 20176981 Free PMC article. - OsWRKY62 and OsWRKY76 Interact with Importin α1s for Negative Regulation of Defensive Responses in Rice Nucleus.
Xu X, Wang H, Liu J, Han S, Lin M, Guo Z, Chen X. Xu X, et al. Rice (N Y). 2022 Feb 20;15(1):12. doi: 10.1186/s12284-022-00558-4. Rice (N Y). 2022. PMID: 35184252 Free PMC article. - Nuclear pore complex composition: a new regulator of tissue-specific and developmental functions.
Raices M, D'Angelo MA. Raices M, et al. Nat Rev Mol Cell Biol. 2012 Nov;13(11):687-99. doi: 10.1038/nrm3461. Nat Rev Mol Cell Biol. 2012. PMID: 23090414 Review. - Quantitative phosphoproteomics identifies SnRK2 protein kinase substrates and reveals the effectors of abscisic acid action.
Wang P, Xue L, Batelli G, Lee S, Hou YJ, Van Oosten MJ, Zhang H, Tao WA, Zhu JK. Wang P, et al. Proc Natl Acad Sci U S A. 2013 Jul 2;110(27):11205-10. doi: 10.1073/pnas.1308974110. Epub 2013 Jun 17. Proc Natl Acad Sci U S A. 2013. PMID: 23776212 Free PMC article.
References
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301, 653–657. - PubMed
- Austin, M.J., Muskett, P., Kahn, K., Feys, B.J., Jones, J.D., and Parker, J.E. (2002). Regulatory role of SGT1 in early R gene-mediated plant defenses. Science 295, 2077–2080. - PubMed
- Azevedo, C., Sadanandom, A., Kitagawa, K., Freialdenhoven, A., Shirasu, K., and Schulze-Lefert, P. (2002). The RAR1 interactor SGT1, an essential component of R gene-triggered disease resistance. Science 295, 2073–2076. - PubMed
- Belkhadir, Y., Nimchuk, Z., Hubert, D.A., Mackey, D., and Dangl, J.L. (2004. a). Arabidopsis RIN4 negatively regulates disease resistance mediated by RPS2 and RPM1 downstream or independent of the NDR1 signal modulator and is not required for the virulence functions of bacterial type III effectors AvrRpt2 or AvrRpm1. Plant Cell 16, 2822–2835. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous