SarA is an essential positive regulator of Staphylococcus epidermidis biofilm development - PubMed (original) (raw)
SarA is an essential positive regulator of Staphylococcus epidermidis biofilm development
María Angeles Tormo et al. J Bacteriol. 2005 Apr.
Abstract
Staphylococcus epidermidis biofilm formation is associated with the production of the polysaccharide intercellular adhesin (PIA)--poly-N-acetylglucosamine polysaccharide (PNAG) by the products of the icaADBC operon. Recent evidence indicates that SarA, a central regulatory element that controls the production of Staphylococcus aureus virulence factors, is essential for the synthesis of PIA/PNAG and the ensuing biofilm development in this species. Based on the presence of a sarA homolog, we hypothesized that SarA could also be involved in the regulation of the biofilm formation process in S. epidermidis. To investigate this, we constructed nonpolar sarA deletions in two genetically unrelated S. epidermidis clinical strains, O-47 and CH845. The SarA mutants were completely defective in biofilm formation, both in the steady-state conditions of a microtiter dish assay and in the flow conditions of microfermentors. Reverse transcription-PCR experiments showed that the mutation in the sarA gene resulted in downregulation of the icaADBC operon transcription in an IcaR-independent manner. Purified SarA protein showed high-affinity binding to the icaA promoter region by electrophoretic mobility shift assays. Consequently, mutation in sarA provoked a significant decrease in the amount of PIA/PNAG on the cell surface. Furthermore, heterologous complementation of S. aureus sarA mutants with the sarA gene of S. epidermidis completely restored biofilm formation. In summary, SarA appeared to be a positive regulator of transcription of the ica locus, and in its absence, PIA/PNAG production and biofilm formation were diminished. Additionally, we present experimental evidence showing that SarA may be an important regulatory element that controls S. epidermidis virulence factors other than biofilm formation.
Figures
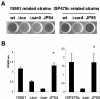
FIG. 1.
Biofilm formation phenotype of two unrelated S. aureus sarA mutant clones carrying pJP19, a shuttle plasmid containing the sarA gene from S. epidermidis. Biofilm formation capacity differences correspond to 24-h biofilm formed on polystyrene microtiter plates after staining with 0.1% safranin. The microtiter plates and mean optical density values obtained (A_495) are shown. (A) Left: wells corresponding to wild-type 15981, 15981 Δ_ica (negative control), 15981 Δ_sarA_, and JP54 (15981 Δ_sarA_ carrying pJP19). Right: wells corresponding to wild-type ISP479c, ISP479c Δ_sarA_, and JP55 (ISP479c Δ_sarA_ carrying pJP19). (B) Mean optical density values. Bars represent the mean values, and error bars represent the standard error of the mean. Significant differences in adherence were noted between complemented and noncomplemented sarA mutant strains (*, P < 0.01).
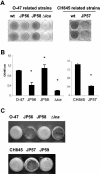
FIG. 2.
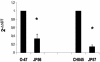
Loss of biofilm formation in two genetically unrelated S. epidermidis sarA mutants. (A) Biofilm formation capacity of S. epidermidis O-47 and CH845 (wild-type strains), their corresponding sarA mutants JP56 and JP57, the JP56 strain complemented with plasmid pJP19 (JP58), and O-47 Δ_ica_::tet as a negative control on polystyrene microtiter plates after 24 h in TSB-gluc medium at 37°C. The bacterial cells were stained with safranin and quantified by determining the absorbance at 495 nm. (B) Significant differences in adherence were noted between wild-type strains and their isogenic sarA mutants as well as between the complemented versus noncomplemented JP56 (O-47 sarA mutant) strain (*, P < 0.05). (C) Phenotypic differences in the capacity to form a 24-h biofilm on the surface of a glass container (visual observation) between wild-type strains O-47 and CH845, their corresponding sarA mutants (JP56 and JP57, respectively), and their sarA mutants complemented with plasmid pJP19 (JP58 and JP59, respectively).
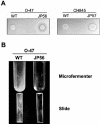
FIG. 3.
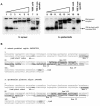
(A) Increased protease production by S. epidermidis sarA mutants. S. epidermidis wild-type strains O-47 and CH845 and their corresponding sarA mutants were grown in skimmed milk agar plates. (B) Biofilm formation in continuous-flow culture microfermenters of S. epidermidis O-47 and its derivative sarA mutant JP56. Biofilm development in microfermenters (upper) or on the corresponding Pyrex slides removed from the microfermenters (lower) after 24 h of growing in TSB-gluc at 37°C is shown.
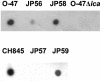
FIG. 4.
Loss of PNAG production in S. epidermidis sarA mutant strains. Cell surface extracts from overnight cultures of S. epidermidis wild-type strains O-47 and CH845, their corresponding sarA mutants JP56 and JP57, respectively, the JP56 and JP57 strains complemented with plasmid pJP19 (strains JP58 and JP 59, respectively), and the O-47 Δ_ica_::tet as a negative control, treated as described in Materials and Methods, were spotted onto nitrocellulose filters. PNAG production was detected with an anti-PNAG polyclonal antibody. The sarA mutants produced lower levels of PNAG product.
FIG. 5.
Real time quantification of ica expression on S. epidermidis wild-type strains and their corresponding sarA mutants. Asterisks denote significance (P < 0.05).
FIG. 6.
Autoradiogram of a nondenaturing 8% polyacrylamide gel with purified SarA protein and a 198-bp γ-32P-radiolabeled DNA fragment containing the intergenic promoter region of the icaRA genes. A. Lanes 1 to 6, mobility of the 198-bp radiolabeled DNA fragment (≈3 ng) of the S. aureus icaRA promoters region in the presence of 0, 50, 100, 200, 300, and 500 ng of purified protein, respectively; lanes 7 and 8, mobility of the same fragment with 300 ng of SarA, but in the presence of a 100-fold excess (molar ratio) of the unlabeled 198-bp fragment as the specific competitor (lane 7) and a 100-fold excess of unlabeled 148-bp intergenic sarUT promoter fragment (36) as the nonspecific competitor (lane 8). Lanes 9 to 16 are similar to lanes 1 to 8 except that the 198-bp fragment was from the ica promoter region of S. epidermidis. B. The nucleotide sequence of the 198-bp fragment containing the ica intergenic region of S. aureus (AF086783) and S. epidermidis (U43366) is shown and marked with the putative binding regions for SarA protein as determined based on the SarA consensus binding site (6).
FIG. 7.
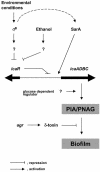
Model of regulation of PIA/PNAG synthesis in S. epidermidis.
Similar articles
- AraC-Type Regulator Rbf Controls the Staphylococcus epidermidis Biofilm Phenotype by Negatively Regulating the icaADBC Repressor SarR.
Rowe SE, Campbell C, Lowry C, O'Donnell ST, Olson ME, Lindgren JK, Waters EM, Fey PD, O'Gara JP. Rowe SE, et al. J Bacteriol. 2016 Oct 7;198(21):2914-2924. doi: 10.1128/JB.00374-16. Print 2016 Nov 1. J Bacteriol. 2016. PMID: 27501984 Free PMC article. - SigmaB and SarA independently regulate polysaccharide intercellular adhesin production in Staphylococcus epidermidis.
Handke LD, Slater SR, Conlon KM, O'Donnell ST, Olson ME, Bryant KA, Rupp ME, O'Gara JP, Fey PD. Handke LD, et al. Can J Microbiol. 2007 Jan;53(1):82-91. doi: 10.1139/w06-108. Can J Microbiol. 2007. PMID: 17496953 - SarA and not sigmaB is essential for biofilm development by Staphylococcus aureus.
Valle J, Toledo-Arana A, Berasain C, Ghigo JM, Amorena B, Penadés JR, Lasa I. Valle J, et al. Mol Microbiol. 2003 May;48(4):1075-87. doi: 10.1046/j.1365-2958.2003.03493.x. Mol Microbiol. 2003. PMID: 12753197 - Polysaccharide intercellular adhesin in biofilm: structural and regulatory aspects.
Arciola CR, Campoccia D, Ravaioli S, Montanaro L. Arciola CR, et al. Front Cell Infect Microbiol. 2015 Feb 10;5:7. doi: 10.3389/fcimb.2015.00007. eCollection 2015. Front Cell Infect Microbiol. 2015. PMID: 25713785 Free PMC article. Review. - Genetic regulation of the intercellular adhesion locus in staphylococci.
Cue D, Lei MG, Lee CY. Cue D, et al. Front Cell Infect Microbiol. 2012 Mar 26;2:38. doi: 10.3389/fcimb.2012.00038. eCollection 2012. Front Cell Infect Microbiol. 2012. PMID: 23061050 Free PMC article. Review.
Cited by
- The two-component signal transduction system ArlRS regulates Staphylococcus epidermidis biofilm formation in an ica-dependent manner.
Wu Y, Wang J, Xu T, Liu J, Yu W, Lou Q, Zhu T, He N, Ben H, Hu J, Götz F, Qu D. Wu Y, et al. PLoS One. 2012;7(7):e40041. doi: 10.1371/journal.pone.0040041. Epub 2012 Jul 27. PLoS One. 2012. PMID: 22848368 Free PMC article. - Staphylococcus aureus develops an alternative, ica-independent biofilm in the absence of the arlRS two-component system.
Toledo-Arana A, Merino N, Vergara-Irigaray M, Débarbouillé M, Penadés JR, Lasa I. Toledo-Arana A, et al. J Bacteriol. 2005 Aug;187(15):5318-29. doi: 10.1128/JB.187.15.5318-5329.2005. J Bacteriol. 2005. PMID: 16030226 Free PMC article. - Flavaspidic acid BB combined with mupirocin improves its anti-bacterial and anti-biofilm activities against Staphylococcus epidermidis.
Cai Z, Mo Z, Zheng S, Lan S, Xie S, Lu J, Tang C, Shen Z. Cai Z, et al. BMC Microbiol. 2022 Jul 15;22(1):179. doi: 10.1186/s12866-022-02578-y. BMC Microbiol. 2022. PMID: 35840879 Free PMC article. - Staphylococcal biofilms.
Otto M. Otto M. Curr Top Microbiol Immunol. 2008;322:207-28. doi: 10.1007/978-3-540-75418-3_10. Curr Top Microbiol Immunol. 2008. PMID: 18453278 Free PMC article. Review. - Virulence Factors in Coagulase-Negative Staphylococci.
França A, Gaio V, Lopes N, Melo LDR. França A, et al. Pathogens. 2021 Feb 4;10(2):170. doi: 10.3390/pathogens10020170. Pathogens. 2021. PMID: 33557202 Free PMC article. Review.
References
- Augustin, J., R. Rosenstein, B. Wieland, U. Schneider, N. Schnell, G. Engelke, K. D. Entian, and F. Götz. 1992. Genetic analysis of epidermin biosynthetic genes and epidermin-negative mutants of Staphylococcus epidermidis. Eur. J. Biochem. 204:1149-1154. - PubMed
- Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1990. Current protocols in molecular biology. John Wiley & Sons, New York, N.Y.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous