Role of oxidative carbonylation in protein quality control and senescence - PubMed (original) (raw)
Review
Role of oxidative carbonylation in protein quality control and senescence
Thomas Nyström. EMBO J. 2005.
Abstract
Proteins can become modified by a large number of reactions involving reactive oxygen species. Among these reactions, carbonylation has attracted a great deal of attention due to its irreversible and unrepairable nature. Carbonylated proteins are marked for proteolysis by the proteasome and the Lon protease but can escape degradation and form high-molecular-weight aggregates that accumulate with age. Such carbonylated aggregates can become cytotoxic and have been associated with a large number of age-related disorders, including Parkinson's disease, Alzheimer's disease, and cancer. This review focuses on the generation of and defence against protein carbonyls and speculates on the potential role of carbonylation in protein quality control, cellular deterioration, and senescence.
Figures
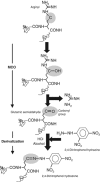
Figure 1
Carbonylation and derivatization of a protein amino-acid side chain. A scheme for the formation of glutamic semialdehyde from an arginyl residue is depicted as a consequence of an MCO. For detection, the carbonyl group, in this case glutamic semialdehyde, is subsequently derivatized by 2,4-dinitrophenol hydrazine. The resulting protein 2,4-dinitrophenol hydrazone can be detected by specific monoclonal or polyclonal antibodies (see Requena et al, 2001).
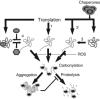
Figure 2
Schematic representation of pathways that can generate carbonylation-prone protein species and the fate of such proteins. Pathway 1 portrays the generation of aberrant proteins by mistranslation, for example, missense incorporation, frame-shifting, and stop-codon read-through. Pathway 2 signifies the formation of aberrant folding structures during chaperone deficiency, whereas pathway 3 is the formation of similar aberrant structures during specific stresses such as heat and exposure to denaturing agents. Pathway 4 illustrates the hypothetical carbonylation of an idle enzyme structure, which prevails during substrate deficiency. As long as the substrate is present and the enzyme working, it may be safeguarded from carbonylation and less prone to be attacked by proteases. Soluble species of carbonylated proteins appear ordained for proteolysis by, for example, the proteasome or Lon protease. However, highly carbonylated proteins form high-molecular-weight aggregates that are proteolysis-resistant. Such aggregates appear to inhibit protease functions.
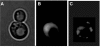
Figure 3
Segregation of carbonylated proteins during cytokinesis of the yeast S. cerevisiae. (A) Transmission electrograph of a yeast mother cell in the process of generating a daughter cell. (B) Levels of superoxide as determined by DHE staining in cells shown in panel A. (C) In situ detection of carbonylated proteins (see Aguilaniu et al, 2003) of cells shown in panel A.
Similar articles
- Pattern of occurrence and occupancy of carbonylation sites in proteins.
Rao RS, Møller IM. Rao RS, et al. Proteomics. 2011 Nov;11(21):4166-73. doi: 10.1002/pmic.201100223. Epub 2011 Sep 14. Proteomics. 2011. PMID: 21919202 - Accumulation of altered proteins and ageing: causes and effects.
Hipkiss AR. Hipkiss AR. Exp Gerontol. 2006 May;41(5):464-73. doi: 10.1016/j.exger.2006.03.004. Epub 2006 Apr 18. Exp Gerontol. 2006. PMID: 16621390 Review. - Proteome-wide profiling of carbonylated proteins and carbonylation sites in HeLa cells under mild oxidative stress conditions.
Bollineni RC, Hoffmann R, Fedorova M. Bollineni RC, et al. Free Radic Biol Med. 2014 Mar;68:186-95. doi: 10.1016/j.freeradbiomed.2013.11.030. Epub 2013 Dec 7. Free Radic Biol Med. 2014. PMID: 24321318 - Oxidative proteome modifications target specific cellular pathways during oxidative stress, cellular senescence and aging.
Baraibar MA, Friguet B. Baraibar MA, et al. Exp Gerontol. 2013 Jul;48(7):620-5. doi: 10.1016/j.exger.2012.10.007. Epub 2012 Nov 2. Exp Gerontol. 2013. PMID: 23127722 Review. - Mitochondrial protein quality control: implications in ageing.
Friguet B, Bulteau AL, Petropoulos I. Friguet B, et al. Biotechnol J. 2008 Jun;3(6):757-64. doi: 10.1002/biot.200800041. Biotechnol J. 2008. PMID: 18446870 Review.
Cited by
- Effects of Ramadan Fasting on Serum Amyloid A and Protein Carbonyl Group Levels in Patients With Cardiovascular Diseases.
Asadi H, Abolfathi AA, Badalzadeh R, Majidinia M, Yaghoubi A, Asadi M, Yousefi B. Asadi H, et al. J Cardiovasc Thorac Res. 2015;7(2):55-9. doi: 10.15171/jcvtr.2015.12. J Cardiovasc Thorac Res. 2015. PMID: 26191392 Free PMC article. - Physicochemical and biological impact of metal-catalyzed oxidation of IgG1 monoclonal antibodies and antibody-drug conjugates via reactive oxygen species.
Glover ZK, Wecksler A, Aryal B, Mehta S, Pegues M, Chan W, Lehtimaki M, Luo A, Sreedhara A, Rao VA. Glover ZK, et al. MAbs. 2022 Jan-Dec;14(1):2122957. doi: 10.1080/19420862.2022.2122957. MAbs. 2022. PMID: 36151884 Free PMC article. - Shotgun redox proteomics: identification and quantitation of carbonylated proteins in the UVB-resistant marine bacterium, Photobacterium angustum S14.
Matallana-Surget S, Cavicchioli R, Fauconnier C, Wattiez R, Leroy B, Joux F, Raftery MJ, Lebaron P. Matallana-Surget S, et al. PLoS One. 2013 Jul 9;8(7):e68112. doi: 10.1371/journal.pone.0068112. Print 2013. PLoS One. 2013. PMID: 23874515 Free PMC article. - DNA-PK Deficiency in Alzheimer's Disease.
Kanungo J. Kanungo J. J Neurol Neuromedicine. 2016 Sep;1(3):17-22. doi: 10.29245/2572.942x/2016/3.1016. J Neurol Neuromedicine. 2016. PMID: 27376156 Free PMC article. - Chemical and biological methods to detect post-translational modifications of arginine.
Slade DJ, Subramanian V, Fuhrmann J, Thompson PR. Slade DJ, et al. Biopolymers. 2014 Feb;101(2):133-43. doi: 10.1002/bip.22256. Biopolymers. 2014. PMID: 23576281 Free PMC article. Review.
References
- Agarwal S, Sohal RS (1994) Aging and proteolysis of oxidized proteins. Arch Biochem Biophys 309: 24–28 - PubMed
- Aguilaniu H, Gustafsson L, Rigoulet M, Nyström T (2001) Protein oxidation in G0 cells of Saccharomyces cerevisiae depends on the state rather than rate of respiration and is enhanced in pos9 but not yap1 mutants. J Biol Chem 276: 35396–35404 - PubMed
- Aguilaniu H, Gustafsson L, Rigoulet M, Nyström T (2003) Asymmetric inheritance of oxidatively damaged proteins during cytokinesis in Saccharomyces cerevisiae: a Sir2p dependent mechanism. Science 299: 1751–1753 - PubMed
- Barak Z, Gallant J, Lindsley D, Kwieciszewki B, Heidel D (1996) Enhanced ribosome frameshifting in stationary phase cells. J Mol Biol 263: 140–148 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical