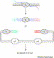Seamless cloning and gene fusion - PubMed (original) (raw)
Review
Seamless cloning and gene fusion
Quinn Lu. Trends Biotechnol. 2005 Apr.
Abstract
Gene fusion technology is a key tool in facilitating gene function studies. Hybrid molecules in which all the components are joined precisely, without the presence of intervening and unwanted extraneous sequences, enable accurate studies of molecules and the characterization of individual components. This article reviews situations in which seamlessly fused genes and proteins are required or desired and describes molecular approaches that are available for generating these hybrid molecules.
Figures
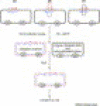
Figure 1
Seamless gene fusion by overlap PCR. The diagram shows seamless fusion of DNA fragments X and Y. The two DNA fragments are PCR amplified individually. Primers P2 and P3 are designed so that the 5′-end 15 bases are complementary to each other. The PCR products are then used as templates for a second PCR amplification with primers P1 and P4. The complementary part of P2 and P3 could be part of fragment X or fragment Y. Note that to facilitate efficient PCR amplification, the melting temperature (_T_m) for all primers should be made to be similar within the range of 55°C–75°C.
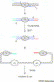
Figure 2
Seamless DNA manipulation by QuickChange™ site-directed mutagenesis. The diagram shows steps involved in site-directed mutagenesis for generating point mutations (a), insertions (b) or deletions (c). In all the cases, two complementary mutagenic primers (or megaprimers in case b) are used with each having >15 base homologous sequences flanking the mutagenic site. After primer extension cycles with Pfu polymerase, the undesired methylated template DNA and semi-methylated hybrids are fragmented by treating with restriction enzyme _Dpn_I. The desired mutant circular duplexes are recovered in E. coli following transformation. The plasmid backbone contains an origin of replication (ori) and a selectable marker (sm).
Figure 3
Seamless gene cloning and gene fusion via a type IIS restriction enzyme. _Sap_I is used as an example. (a) Seamless assembly of fragments X, Y, and Z. The fragments are first individually PCR amplified, with primers containing a _Sap_I site. The primers are so designed that upon _Sap_I digestion specific cohesive ends are generated for each fragment. Following _Sap_I digestion, ligation of the fragments results in seamless assembly of X, Y, and Z. The 5′-end of X and the 3′-end of Z are made to contain _Sap_I (with ends incompatible to other ends of X, Y and Z) or any other restriction sites for further subcloning. (b) Seamless fusion of fragment X with fragments Y and Z contained in a vector. Fragment A is PCR amplified with primers containing a _Sap_I site. The primers are so designed that upon digestion with _Sap_I cohesive ends are generated. The vector fragment, which contains fragments Y and Z, was specially engineered and prepared by _Sap_I digestion. Ligation of the _Sap_I-treated fragment A with the vector fragment results in seamless fusion of X with Y and Z. The plasmid backbone contains an origin of replication (ori) and a selectable marker (sm). Note that the nucleotide bases comprising the cohesive ends can be part of the fragment X or its fusion partners, and can be chosen to make ligation of the fragments directional.
Figure 4
Seamless cloning by ligation-independent cloning (LIC). The diagram shows seamless fusion of fragment X with fragments Y and Z contained in a vector. Fragment X is first PCR amplified and purified. The primers are designed so that the 5′-end 12 bases are free of one specific nucleotide (e.g. dT as an example). The product is then treated with a DNA polymerase possessing 3′-5′ exonuclease activity (such as T4 DNA polymerase and Pfu) in the presence of dATP. The polymerase starts to remove nucleotides from 3′-ends of the fragment until a dA base is encountered and removed, which is subsequently added back by the enzyme's 5′-3′ polymerase activity. This reaction generates 12 base (or longer) overhangs on fragment X. The vector fragment, which contains fragments Y and Z was engineered and prepared similarly to yield 12 base (or longer) overhangs that are complementary to the insert fragment. The plasmid backbone contains an origin of replication (ori) and a selectable marker (sm). The LIC-ready vector fragment and the insert fragment are then annealed to form circular duplexes, which are recovered in E. coli following transformation , . Seamless fusion of X with Y and Z is achieved if the nucleotides comprising the LIC overhangs are made to be parts of X or its fusion partners.
Figure 5
Seamless cloning by in vivo recombination. The diagram shows seamless fusion of fragment X with fragments Y and/or Z contained in a vector. Both the insert and the vector fragments are PCR amplified. The primers used in the reaction are designed so that the products contain a stretch of homologous sequences (15–40 bps) at the ends. The plasmid backbone contains an origin of replication (ori) and a selectable marker (sm). The two DNA fragments are then co-transformed into E. coli for in vivo recombination. Seamless fusion is achieved as long as the homologous sequences are parts of fragment X or its fusion partners.
Similar articles
- A simple and universal ligation mediated fusion of genes based on hetero-staggered PCR for generating immunodominant chimeric proteins.
Reddy PK, Ramlal S, Sripathy MH, Batra H. Reddy PK, et al. Gene. 2012 Nov 1;509(1):104-9. doi: 10.1016/j.gene.2012.08.011. Epub 2012 Aug 16. Gene. 2012. PMID: 22917676 - The all purpose gene fusion.
Beckwith J. Beckwith J. Methods Enzymol. 2000;326:3-7. doi: 10.1016/s0076-6879(00)26043-6. Methods Enzymol. 2000. PMID: 11036631 Review. No abstract available. - Fusion PCR, a one-step variant of the "megaprimer" method of mutagenesis.
Karreman C. Karreman C. Biotechniques. 1998 May;24(5):736, 740, 742. doi: 10.2144/98245bm08. Biotechniques. 1998. PMID: 9591118 No abstract available. - Alternative Seamless Cloning Strategies in Fusing Gene Fragments Based on Overlap-PCR.
Hou XW, Tong HY, He ZH. Hou XW, et al. Mol Biotechnol. 2021 Mar;63(3):221-231. doi: 10.1007/s12033-020-00298-0. Epub 2021 Jan 13. Mol Biotechnol. 2021. PMID: 33439452 - Insertional gene fusion technology.
Doi N, Yanagawa H. Doi N, et al. FEBS Lett. 1999 Aug 20;457(1):1-4. doi: 10.1016/s0014-5793(99)00991-6. FEBS Lett. 1999. PMID: 10486551 Review.
Cited by
- Rationally designed logic integration of regulatory signals in mammalian cells.
Leisner M, Bleris L, Lohmueller J, Xie Z, Benenson Y. Leisner M, et al. Nat Nanotechnol. 2010 Sep;5(9):666-70. doi: 10.1038/nnano.2010.135. Epub 2010 Jul 11. Nat Nanotechnol. 2010. PMID: 20622866 Free PMC article. - Restriction enzyme body doubles and PCR cloning: on the general use of type IIs restriction enzymes for cloning.
Tóth E, Huszár K, Bencsura P, Kulcsár PI, Vodicska B, Nyeste A, Welker Z, Tóth S, Welker E. Tóth E, et al. PLoS One. 2014 Mar 11;9(3):e90896. doi: 10.1371/journal.pone.0090896. eCollection 2014. PLoS One. 2014. PMID: 24618593 Free PMC article. - Automated seamless DNA co-transformation cloning with direct expression vectors applying positive or negative insert selection.
Olieric N, Kuchen M, Wagen S, Sauter M, Crone S, Edmondson S, Frey D, Ostermeier C, Steinmetz MO, Jaussi R. Olieric N, et al. BMC Biotechnol. 2010 Aug 9;10:56. doi: 10.1186/1472-6750-10-56. BMC Biotechnol. 2010. PMID: 20691119 Free PMC article. - Construction and evaluation of a novel bifunctional phenylalanine-formate dehydrogenase fusion protein for bienzyme system with cofactor regeneration.
Jiang W, Fang BS. Jiang W, et al. J Ind Microbiol Biotechnol. 2016 May;43(5):577-84. doi: 10.1007/s10295-016-1738-6. Epub 2016 Jan 27. J Ind Microbiol Biotechnol. 2016. PMID: 26819086 - OEPR Cloning: an Efficient and Seamless Cloning Strategy for Large- and Multi-Fragments.
Liu CJ, Jiang H, Wu L, Zhu LY, Meng E, Zhang DY. Liu CJ, et al. Sci Rep. 2017 Mar 16;7:44648. doi: 10.1038/srep44648. Sci Rep. 2017. PMID: 28300166 Free PMC article.
References
- Lu Q. Plasmid vectors for gene cloning and expression. In: Funnell B.E., Phillips G.J., editors. Plasmid Biology. ASM Press; 2004. pp. 545–566.
- McKnight S.L., Kingsbury R. Transcriptional control signals of a eukaryotic protein-coding gene. Science. 1982;217:316–324. - PubMed
- Patten P.A. Applications of DNA shuffling to pharmaceuticals and vaccines. Curr. Opin. Biotechnol. 1997;8:724–733. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources