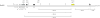Formation of linear inverted repeat amplicons following targeting of an essential gene in Leishmania - PubMed (original) (raw)
Formation of linear inverted repeat amplicons following targeting of an essential gene in Leishmania
Paul-André Genest et al. Nucleic Acids Res. 2005.
Abstract
Attempts to inactivate an essential gene in the protozoan parasite Leishmania have often led to the generation of extra copies of the wild-type alleles of the gene. In experiments with Leishmania tarentolae set up to disrupt the gene encoding the J-binding protein 1 (JBP1), a protein binding to the unusual base beta-D-glucosyl-hydroxymethyluracil (J) of Leishmania, we obtained JBP1 mutants containing linear DNA elements (amplicons) of approximately 100 kb. These amplicons consist of a long inverted repeat with telomeric repeats at both ends and contain either the two different targeting cassettes used to inactivate JBP1, or one cassette and one JBP1 gene. Each long repeat within the linear amplicons corresponds to sequences covering the JBP1 locus, starting at the telomeres upstream of JBP1 and ending in a approximately 220 bp sequence repeated in an inverted (palindromic) orientation downstream of the JBP1 locus. We propose that these amplicons have arisen by a template switch inside a DNA replication fork involving the inverted DNA repeats and helped by the gene targeting.
Figures
Figure 1
Targeting of the _JBP_1 locus and analysis of the mutant cell lines obtained. (A) Map of the JBP1 locus before and after integration of the inactivation cassettes. Sequences flanking the JBP1 locus were cloned upstream and downstream of selection markers. Sequences Y (polypyrimidine stretch) (30) and TUB (intergenic region of the alpha tubulin array of Leishmania enriettii) (49) were used for mRNA processing. The location of the NcoI sites and the JBP1 3′-UTR probe are depicted. The drawing is approximately to scale. (B) Southern blot of genomic DNA digested with NcoI. The blot was hybridized with the JBP1 3′-UTR probe. Lanes: 1, TarIIWT; 2, JBP1 KO HYG; 3, JBP1 KO PUR; 4, JBP1 KO HYG:KO PUR; 5, JBP1 KO NEO; 6, JBP1 KO HYG:KO NEO; 7, JBP1 KO PUR:KO HYG; and 8, JBP1 KO HYG:KO PUR:KO NEO. The alleles present in the different strains are indicated above the lanes. The asterisk identifies the amplified alleles. The wild-type 4.7 kb band can be used as a loading control, except in lane 5 where the loading can be checked by comparison of the amplified 2.7 kb band with lane 6.
Figure 2
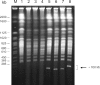
Pulsed-field gel stained with ethidium bromide showing separation of chromosome-sized DNA of L.tarentolae and the amplicon bands present in the JBP1 mutants. The arrow is pointing at the region containing the linear amplicons. Lanes M: Saccharomyces cerevisiae marker; 1, TarIIWT; 2, JBP1 KO HYG; 3, JBP1 KO PUR; 4, JBP1 KO HYG:KO PUR; 5, JBP1 KO NEO; 6, JBP1 KO HYG:KO NEO; 7, JBP1 KO PUR:KO HYG; and 8, JBP1 KO HYG:KO PUR:KO NEO.
Figure 3
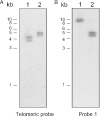
Analysis of the telomeric ends of the JBP1 KO NEO amplicon. Southern blot on digested JBP1 KO NEO amplicon isolated from the pulsed-field gel. The blots were hybridized with a telomeric probe (A) and with probe 1 (see Figure 4), located ∼13 kb upstream of the JBP1 locus (B). Lane 1, BamHI; and lane 2, EcoRV.
Figure 4
Schematic representation of the results of the hybridization experiments done to detect the rearrangement point in the JBP1 KO NEO amplicon. The small rectangles represent the approximate location in the JBP1 chromosome (Chr 9) of the different probes that were generated by PCR on genomic DNA of L.major Friedlin. The white rectangles correspond to the probes that are not present in the amplicon, the black rectangles depict the probes that are present in the amplicon. The gray rectangles correspond to the probes that detect the rearrangement point. The bars indicate the approximate locations of the EcoRV sites around the JBP1 locus in L.tarentolae. The small arrows indicate the location of the RS1, RS2 and RS3 repeats. Their orientation is indicated by the direction of the arrow. The triangles represent the telomeric repeats. The JBP1 gene is depicted by the yellow rectangle. The drawing is approximately to scale.
Figure 5
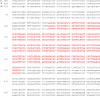
Sequence of the rearrangement point of the JBP1 KO NEO amplicon. Sequence alignment of the inversion point of the JBP1 KO NEO amplicon with the corresponding sequence in the JBP1 wild-type chromosome (i.e. sequences bordering the RS2 and RS3 repeats). The red capital letters depict the perfect homology between the inversion point of the amplicon (KO NEO line) and the RS2 and RS3 repeats. The capital letters show the homology between the sequence surrounding the rearrangement point and the corresponding sequences on the JBP1 chromosome. The orientation of the sequence is the one used in the bottom panel of Figure 6A, where the 5′ end of the sequence corresponds to the left-hand side of the schematic (i.e. the RS2 sequence is presented in an inverted way compared to the wild-type orientation). Note the presence of a small mismatch inside the homologous sequence of RS2 and RS3. We have not determined whether this is a polymorphism between RS2 and RS3 or a point mutation introduced during our cloning procedure.
Figure 6
Structure of the JBP1 KO NEO amplicon. (A) Schematic representation of the right end of chromosome 9 of L.tarentolae (top panel) and of the JBP1 KO NEO amplicon (bottom). The amplicon is one long inverted repeat (depicted by the long broken arrows) with an RS repeat located at the rearrangement point (shown by the yellow star). The location of the A, B, C and D primers and their orientation is shown by the colored triangles. The small black triangles correspond to the telomeres. The RS repeats are depicted by the small arrows, the JBP1 and NEO genes by the colored boxes. We have not determined on which arms each gene lays. (B) PCR products obtained using two primers (A and B) oriented in the same way towards the RS2 and RS3 repeats (left panel) or using a combination of primers (C and D) located on both sides of the RS3 repeat (right panel). Lanes: 1, Marker; 2, TarIIWT; 3, JBP1 KO NEO; 4, JBP1 KO NEO revertant; 5, JBP1 KO NEO purified amplicon; and 6, TarIIWT + JBP1 KO NEO amplicon.
Figure 7
Postulated model to explain the generation of the JBP1 KO NEO amplicon. We propose that replication starts upstream of JBP1. Two copies of JBP1 are generated after the replication fork has passed the JBP1 locus. One of these copies is targeted by the inactivation construct, which replaces JBP1 by NEO (A). The perturbation caused by the targeting could cause a stalling of the replication fork. Reversal of the fork would enable the RS3 repeat of the leading strand to anneal with the not yet replicated RS2 repeat of the lagging strand (B). Replication would restart using the other strand as a template followed by the synthesis of the complementary strand (C). Pealing of the misreplicated section would lead to an amplicon having an inverted repeat structure and two single-stranded DNA having on one strand JBP1 and on the other the NEO marker (D). Annealing of the strands would create a heteroduplex that would be repaired (E). The newly introduced marker would end up in the JBP1 chromosome only in half of the events, depending on which strand is repaired (F). The leading strand of the replication fork is shown in orange, the lagging strand in blue. The RS and telomeric repeats, as well as the JBP1 and NEO genes, are depicted as in Figure 6.
Similar articles
- Homologous recombination between direct repeat sequences yields P-glycoprotein containing amplicons in arsenite resistant Leishmania.
Grondin K, Papadopoulou B, Ouellette M. Grondin K, et al. Nucleic Acids Res. 1993 Apr 25;21(8):1895-901. doi: 10.1093/nar/21.8.1895. Nucleic Acids Res. 1993. PMID: 8098523 Free PMC article. - Defining the sequence requirements for the positioning of base J in DNA using SMRT sequencing.
Genest PA, Baugh L, Taipale A, Zhao W, Jan S, van Luenen HG, Korlach J, Clark T, Luong K, Boitano M, Turner S, Myler PJ, Borst P. Genest PA, et al. Nucleic Acids Res. 2015 Feb 27;43(4):2102-15. doi: 10.1093/nar/gkv095. Epub 2015 Feb 6. Nucleic Acids Res. 2015. PMID: 25662217 Free PMC article. - The domain architecture of the protozoan protein J-DNA-binding protein 1 suggests synergy between base J DNA binding and thymidine hydroxylase activity.
Adamopoulos A, Heidebrecht T, Roosendaal J, Touw WG, Phan IQ, Beijnen J, Perrakis A. Adamopoulos A, et al. J Biol Chem. 2019 Aug 23;294(34):12815-12825. doi: 10.1074/jbc.RA119.007393. Epub 2019 Jul 10. J Biol Chem. 2019. PMID: 31292194 Free PMC article. - Drug resistance and P-glycoprotein gene amplification in the protozoan parasite Leishmania.
Ouellette M, Borst P. Ouellette M, et al. Res Microbiol. 1991 Jul-Aug;142(6):737-46. doi: 10.1016/0923-2508(91)90089-s. Res Microbiol. 1991. PMID: 1961984 Review. - Base J: discovery, biosynthesis, and possible functions.
Borst P, Sabatini R. Borst P, et al. Annu Rev Microbiol. 2008;62:235-51. doi: 10.1146/annurev.micro.62.081307.162750. Annu Rev Microbiol. 2008. PMID: 18729733 Review.
Cited by
- Knockout of protein phosphatase 1 in Leishmania major reveals its role during RNA polymerase II transcription termination.
Kieft R, Zhang Y, Yan H, Schmitz RJ, Sabatini R. Kieft R, et al. Nucleic Acids Res. 2023 Jul 7;51(12):6208-6226. doi: 10.1093/nar/gkad394. Nucleic Acids Res. 2023. PMID: 37194692 Free PMC article. - Behind Base J: The Roles of JBP1 and JBP2 on Trypanosomatids.
Assis LHC, de Paiva SC, Cano MIN. Assis LHC, et al. Pathogens. 2023 Mar 16;12(3):467. doi: 10.3390/pathogens12030467. Pathogens. 2023. PMID: 36986389 Free PMC article. Review. - Genetic Interaction Between Site-Specific Epigenetic Marks and Roles of H4v in Transcription Termination in Trypanosoma brucei.
Kim HS. Kim HS. Front Cell Dev Biol. 2021 Oct 14;9:744878. doi: 10.3389/fcell.2021.744878. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34722526 Free PMC article. - Chromatin-Associated Protein Complexes Link DNA Base J and Transcription Termination in Leishmania.
Jensen BC, Phan IQ, McDonald JR, Sur A, Gillespie MA, Ranish JA, Parsons M, Myler PJ. Jensen BC, et al. mSphere. 2021 Feb 24;6(1):e01204-20. doi: 10.1128/mSphere.01204-20. mSphere. 2021. PMID: 33627513 Free PMC article. - _J_-binding protein 1 and _J_-binding protein 2 expression in clinical Leishmania major no response-antimonial isolates.
Ahmadian S, Eslami G, Fatahi A, Hosseini SS, Vakili M, Ajamein Fahadan V, Elloumi M. Ahmadian S, et al. J Parasit Dis. 2019 Mar;43(1):39-45. doi: 10.1007/s12639-018-1052-5. Epub 2018 Nov 20. J Parasit Dis. 2019. PMID: 30956444 Free PMC article.
References
- Mottram J.C., McCready B.P., Brown K.G., Grant K.M. Gene disruptions indicate an essential function for the LmmCRK1 cdc2-related kinase of Leishmania mexicana. Mol. Microbiol. 1996;22:573–583. - PubMed
- Beverley S.M., Coderre J.A., Santi D.V., Schimke R.T. Unstable DNA amplifications in methotrexate-resistant Leishmania consist of extrachromosomal circles which relocalize during stabilization. Cell. 1984;38:431–439. - PubMed
- White T.C., Fase-Fowler F., van Luenen H., Calafat J., Borst P. The H circles of Leishmania tarentolae are a unique amplifiable system of oligomeric DNAs associated with drug resistance. J. Biol. Chem. 1988;263:16977–16983. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials